Optimized Enzymatic Digestion Protocols for Mesenchymal Stromal Cell Isolation: A Comprehensive Guide for Researchers
This article provides a detailed, evidence-based guide for researchers and drug development professionals on enzymatic digestion protocols for isolating Mesenchymal Stromal Cells (MSCs) from diverse tissue sources.
Optimized Enzymatic Digestion Protocols for Mesenchymal Stromal Cell Isolation: A Comprehensive Guide for Researchers
Abstract
This article provides a detailed, evidence-based guide for researchers and drug development professionals on enzymatic digestion protocols for isolating Mesenchymal Stromal Cells (MSCs) from diverse tissue sources. Covering foundational principles to advanced applications, it explores tissue-specific methodologies for bone marrow, adipose tissue, and umbilical cord, alongside optimization strategies for enzymes like Collagenase and Liberase™. The content further addresses critical troubleshooting, quality control measures per ISCT criteria, and comparative analyses with explant methods. By synthesizing current research and regulatory considerations for Advanced Therapy Medicinal Products (ATMPs), this resource aims to standardize and enhance the efficiency of MSC isolation for robust regenerative medicine applications.
Understanding Mesenchymal Stromal Cells and the Role of Enzymatic Digestion
Historical Context and Nomenclature Evolution
The concept of mesenchymal stromal cells (MSCs) has undergone significant evolution since its initial discovery, reflecting advances in understanding their biological nature and therapeutic mechanisms. The historical foundation of MSCs traces back to pioneering work by Friedenstein and colleagues in the 1960s and 1970s, who first identified adherent, fibroblast-like cells in bone marrow capable of forming discrete colonies (CFU-Fs) and generating multiple skeletal tissues upon transplantation [1]. These cells were initially termed "osteogenic stem cells" or "bone marrow stromal stem cells" [1].
The term "mesenchymal stem cells" (MSCs) was formally coined by Arnold Caplan in 1991, gaining wide popularity following studies demonstrating their multipotent differentiation potential [1] [2] [3]. However, as research progressed, evidence revealed that their therapeutic effects primarily stem from paracrine signaling and immunomodulatory mechanisms rather than lineage-driven tissue regeneration [4]. This biological understanding prompted a nomenclature shift endorsed by the International Society for Cell & Gene Therapy (ISCT), which recommended "mesenchymal stromal cells" in 2019 to better reflect their in vivo properties and primary mechanisms of action [5] [4]. Contemporary literature increasingly frames these interventions as MSC-based immunomodulatory therapies to enhance scientific clarity and align with their clinical applications [4].
Current ISCT Defining Criteria
The ISCT has established minimal criteria for defining human MSCs, which remain fundamental for their identification and characterization in research and clinical applications [5].
- Plastic Adherence: MSCs must adhere to tissue culture plastic under standard culture conditions.
- Specific Surface Marker Expression: MSCs must express specific positive markers and lack expression of negative markers.
- Trilineage Differentiation Potential: MSCs must demonstrate capacity to differentiate into osteoblasts, adipocytes, and chondrocytes under standard in vitro inducing conditions.
Table 1: ISCT Minimal Defining Criteria for Human MSCs
| Criterion | Requirement | Standard Assay |
|---|---|---|
| Plastic Adherence | Adherent under standard culture conditions | Primary cell culture observation |
| Positive Marker Expression | ≥95% positive for CD105, CD73, CD90 | Flow cytometry analysis |
| Negative Marker Expression | ≤2% positive for CD45, CD34, CD14/CD11b, CD79α/CD19, HLA-DR | Flow cytometry analysis |
| Multilineage Differentiation | In vitro differentiation to osteocytes, adipocytes, chondrocytes | Specific staining after culture in induction media |
MSC Isolation Methods and Protocols
MSCs can be isolated from various tissue sources using different methodological approaches. The choice of method depends on the tissue source, intended application, and required cell yield and viability.
- Bone Marrow: The original and most frequently utilized source of MSCs [5].
- Adipose Tissue: Provides higher yield and easier access compared to bone marrow [5] [6].
- Umbilical Cord/Wharton's Jelly: A rich perinatal source with primitive properties and fast growth rate [7] [3] [5].
- Other Sources: Dental pulp, placenta, amniotic fluid, and menstrual blood [3] [5].
Isolation Techniques: Enzymatic Digestion vs. Explant Method
Two primary isolation methods are employed, each with distinct advantages and limitations.
Enzymatic Digestion Protocol
This method uses enzymes to digest the extracellular matrix and release individual cells.
Detailed Protocol for Adipose Tissue-Derived MSCs [6] [8]:
- Tissue Processing: Wash adipose tissue extensively with phosphate-buffered saline (PBS) containing antibiotics (e.g., 100 U/mL penicillin, 0.1 mg/mL streptomycin, 0.25 μg/mL amphotericin B).
- Enzymatic Digestion: Mince tissue and digest with appropriate enzyme solution:
- Enzyme Options: Collagenase type I (0.1-0.4%), Liberase TM (0.1-0.2%), or Collagenase type IV.
- Incubation Conditions: 37°C for 30 minutes to 3 hours with agitation.
- Digestion Neutralization: Add complete culture medium (e.g., Dulbecco's Modified Eagle Medium with 10% fetal bovine serum) to neutralize enzymes.
- Cell Recovery: Centrifuge suspension (1200 rpm for 10 minutes) to obtain stromal vascular fraction pellet.
- Cell Seeding and Culture: Resuspend pellet in culture medium and seed in tissue culture flasks. Maintain at 37°C in 5% CO₂.
- Medium Changes: Replace medium after 24-48 hours to remove non-adherent cells, then refresh twice weekly.
Explant Culture Protocol
This method relies on MSC migration from tissue fragments placed in culture.
Detailed Protocol for Umbilical Cord-Derived MSCs [7] [3]:
- Tissue Preparation: Remove vessels from umbilical cord and wash thoroughly with PBS containing antibiotics.
- Tissue Sectioning: Dissect Wharton's jelly into small segments (2-3 mm diameter).
- Explant Seeding: Place tissue segments in culture dishes without enzymatic treatment.
- Culture Conditions: Maintain explants in low-glucose DMEM with 10% FBS and antibiotics in a humidified 37°C, 5% CO₂ incubator.
- Medium Management: Replace culture media twice weekly.
- Cell Harvesting: Remove tissue segments after 2-3 weeks when adequate MSC outgrowth is observed, and continue culture until confluence.
Diagram Title: MSC Isolation and Characterization Workflow
Comparative Analysis of Isolation Methods
Table 2: Comparison of Enzymatic vs. Explant Isolation Methods
| Parameter | Enzymatic Digestion | Explant Method |
|---|---|---|
| Principle | Chemical breakdown of ECM to release individual cells | Cellular migration from tissue explants |
| Procedure Duration | Shorter active processing (50-210 min) [6] | Longer culture time until cell emergence (10-21 days) [7] [8] |
| Cell Yield | Generally higher immediate yield [7] | Lower initial yield, requires expansion |
| Cell Viability | 70%-99% [6] | Comparable viability |
| Tissue Preservation | Disrupts native tissue architecture | Preserves tissue microenvironment and ECM |
| Growth Factor Release | Lower levels in supernatant | Higher levels of natural growth factors (e.g., bFGF) [7] |
| Reproducibility | Highly reproducible | More variable between operators |
| Cost | Higher (enzyme costs) [6] | Lower (no enzymes required) |
| Regulatory Considerations | Defined process, easier standardization | Process validation more complex |
MSC Characterization and Quality Assessment
Immunophenotyping by Flow Cytometry
Comprehensive immunophenotyping is essential for MSC characterization according to ISCT criteria. The following protocol details standard flow cytometry analysis for MSCs:
Sample Preparation:
- Harvest MSCs at 70-80% confluence using trypsin/EDTA.
- Wash cells twice with PBS containing 2% FBS.
- Aliquot approximately 1×10⁵ cells per tube for antibody staining.
Antibody Staining:
- Incubate cells with fluorochrome-conjugated antibodies for 30 minutes on ice in the dark.
- Positive Marker Panel: CD105-FITC, CD90-PE, CD73-APC.
- Negative Marker Panel: CD45-FITC, CD34-PE, CD14/CD11b-APC, HLA-DR-APC.
- Include appropriate isotype controls for each fluorochrome.
Analysis:
- Analyze samples using flow cytometer with minimum 10,000 events per sample.
- Gate on viable cells based on forward/side scatter properties.
- Determine percentage positive cells compared to isotype controls.
- Validated MSCs must demonstrate ≥95% expression of positive markers and ≤2% expression of negative markers.
Trilineage Differentiation Potential
Functional validation of MSC multipotency requires demonstration of differentiation into mesodermal lineages.
Adipogenic Differentiation Protocol:
- Culture MSCs to confluence in growth medium.
- Induce with adipogenic medium (DMEM with 10% FBS, 1 μM dexamethasone, 0.5 mM IBMX, 10 μg/mL insulin, 200 μM indomethacin).
- Maintain for 2-3 weeks with medium changes every 3-4 days.
- Confirm differentiation by Oil Red O staining of lipid vacuoles.
Osteogenic Differentiation Protocol:
- Culture MSCs at 70% confluence in growth medium.
- Induce with osteogenic medium (DMEM with 10% FBS, 0.1 μM dexamethasone, 10 mM β-glycerophosphate, 50 μM ascorbate-2-phosphate).
- Maintain for 3-4 weeks with medium changes twice weekly.
- Confirm differentiation by Alizarin Red S staining of calcium deposits.
Chondrogenic Differentiation Protocol:
- Pellet 2.5×10⁵ MSCs by centrifugation in conical tube.
- Induce with chondrogenic medium (DMEM with 1% ITS+1, 0.1 μM dexamethasone, 50 μM ascorbate-2-phosphate, 40 μg/mL proline, 10 ng/mL TGF-β3).
- Maintain for 3-4 weeks with medium changes every 3-4 days.
- Confirm differentiation by Alcian Blue staining of sulfated proteoglycans.
Research Reagent Solutions
Table 3: Essential Reagents for MSC Isolation and Characterization
| Reagent Category | Specific Examples | Function/Purpose |
|---|---|---|
| Digestive Enzymes | Collagenase Type I/II/IV, Liberase TM, Trypsin | ECM degradation for cell isolation [7] [8] |
| Culture Media | Low-glucose DMEM, α-MEM, DMEM/F12 | Cell nutrition and maintenance |
| Serum Supplements | Fetal Bovine Serum (FBS), Human Serum | Growth factors and adhesion factors |
| Antibiotics | Penicillin, Streptomycin, Amphotericin B | Microbial contamination prevention [7] |
| Flow Cytometry Antibodies | CD73, CD90, CD105, CD45, CD34, HLA-DR | Immunophenotype characterization [5] |
| Differentiation Inducers | Dexamethasone, IBMX, Insulin, TGF-β3, Ascorbate-2-phosphate | Lineage-specific differentiation induction |
| Staining Reagents | Oil Red O, Alizarin Red S, Alcian Blue | Differentiation confirmation |
| Cell Detachment Agents | Trypsin/EDTA, Accutase | Cell harvesting for subculture |
Clinical Applications and Regulatory Considerations
MSCs have advanced significantly toward clinical applications, with recent regulatory approvals underscoring their therapeutic potential. The first FDA-approved MSC product, remestemcel-L-rknd (Ryoncil), was authorized for steroid-refractory pediatric acute graft-versus-host disease (aGVHD) in 2024, followed by China's approval of Amimestrocel injection (Ruibosheng) for aGVHD in 2025 [4]. These milestones signal the maturation of MSC-based therapies and highlight the importance of mechanism-aligned terminology framing these interventions as MSC-based immunomodulatory therapies [4].
Clinical trials have expanded to include autoimmune conditions such as Crohn's disease, multiple sclerosis, systemic lupus erythematosus, type 1 diabetes, and rheumatoid arthritis [4]. In these applications, MSCs demonstrate benefits primarily through immunomodulation rather than lineage-driven regeneration, employing multiple mechanisms including suppression of effector T-cell activation, expansion of regulatory T cells, inhibition of dendritic cell maturation, and reprogramming of myeloid cells toward inflammation-resolving phenotypes [4].
For clinical translation, the ISCT emphasizes the need for standardized reporting of critical quality attributes (CQAs) including donor characteristics, tissue source, isolation method, expansion protocol, population doubling levels, and comprehensive characterization data [9]. Appropriate potency assays aligned with the mechanism of action must be implemented, particularly focusing on immunomodulatory potential for inflammation-related applications [9] [4].
Mesenchymal Stromal Cells (MSCs) represent a cornerstone of regenerative medicine and therapeutic applications due to their multipotent differentiation capacity, immunomodulatory properties, and role in tissue homeostasis [10] [3]. The selection of an appropriate tissue source is a critical primary decision that significantly influences isolation efficiency, cell yield, proliferative capacity, and ultimately, the success of downstream applications. Among the various sources available, bone marrow, adipose tissue, and perinatal tissues have emerged as the most prominent and well-characterized [10] [3]. This application note provides a structured comparison of these three major sources, detailing their respective isolation protocols, key characteristics, and experimental considerations to guide researchers in selecting and implementing the most suitable methodology for their specific research context within a broader thesis on enzymatic digestion protocols for MSC isolation.
Source Comparison and Selection
The choice of tissue source dictates the isolation strategy, yield, and fundamental properties of the resulting MSCs. The table below provides a quantitative and qualitative comparison of the three major sources to inform experimental design.
Table 1: Comparative Analysis of Major MSC Tissue Sources
| Characteristic | Bone Marrow (BM) | Adipose Tissue (AT) | Perinatal Tissues (e.g., Umbilical Cord) |
|---|---|---|---|
| Relative Abundance of MSCs | Low (0.001-0.01% of mononuclear cells) [11] | High (500 to 2500 times greater than BM) [12] [11] | Variable, but generally high [3] |
| Typical Cell Yield | Varies with aspiration volume and donor | ~30-130 x 10⁶ cells/g tissue (bovine AT) [8] | Varies with dissection efficiency and cord size |
| Isolation Primary Method | Density gradient centrifugation and/or plastic adherence [13] [14] | Enzymatic digestion (e.g., Collagenase) [12] [8] | Enzymatic digestion or explant culture [3] [15] |
| Invasiveness of Harvest | High (painful aspiration) | Low (minimally invasive lipoaspiration) [11] | Non-invasive (medical waste post-birth) [3] [15] |
| Proliferative Capacity | Moderate, can exhibit senescence [14] | High | High, considered more primitive [3] [15] |
| Key Advantages | Gold standard, well-characterized | High yield, accessible, minimal donor discomfort | Low immunogenicity, no ethical concerns, high proliferative rate [3] [15] |
| Key Limitations | Low yield, invasive harvest, decline with donor age | Presence of contaminating adipocytes | Finite tissue supply, dependent on birth events |
Detailed Isolation Protocols
Bone Marrow Aspirate: Enzymatic Digestion and Density Gradient Protocol
The isolation of MSCs from bone marrow relies on separating mononuclear cells from the bulk of hematopoietic cells and bone spicules.
Materials & Reagents:
- Alpha Minimum Essential Medium (α-MEM) or Dulbecco's Modified Eagle Medium (DMEM) [11]
- Ficoll-Paque Premium or similar density gradient medium [11] [16]
- Phosphate Buffered Saline (PBS) without Ca²⁺/Mg²⁺
- Penicillin-Streptomycin and/or Gentamycin [11]
- Fetal Bovine Serum (FBS), preferably MSC-qualified
- Collagenase Type I or II (for protocols involving bone fragment digestion) [13]
Procedure:
- Collection and Preparation: Collect bone marrow aspirate into a heparinized syringe. Dilute the aspirate 1:1 with PBS [11] [14].
- Density Gradient Centrifugation: Carefully layer the diluted marrow over Ficoll-Paque in a centrifuge tube. Centrifuge at 400 x g for 30 minutes at room temperature with the brake disengaged [16]. The mononuclear cells (MNCs), including MSCs, will form a buffy coat at the sample-medium interface.
- Cell Washing: Transfer the buffy coat layer to a new tube. Add at least 3 volumes of PBS and centrifuge at 400 x g for 10 minutes. Repeat the wash step [16].
- Plating and Culture: Resuspend the final cell pellet in complete culture medium (e.g., α-MEM supplemented with 10-20% FBS and antibiotics). Seed the cells into culture flasks and incubate at 37°C with 5% CO₂ [11] [14].
- Medium Change and Expansion: After 48-72 hours, replace the medium to remove non-adherent hematopoietic cells. Refresh the medium every 3-4 days thereafter. Passage adherent cells upon reaching 70-80% confluence [11].
The workflow for this protocol is standardized as follows:
Adipose Tissue: Optimized Enzymatic Digestion Protocol
Adipose tissue is digested to release the Stromal Vascular Fraction (SVF), which contains AD-MSCs, endothelial cells, and pericytes.
Materials & Reagents:
- Dulbecco's Phosphate Buffered Saline (DPBS)
- Collagenase Type I, Liberase, or other optimized enzyme mixtures [12] [8]
- Complete growth medium (e.g., DMEM with 10% FBS)
- NH₄Cl for red blood cell lysis (optional) [16]
- 40-100µm cell strainers
Procedure:
- Tissue Washing: Mince the adipose tissue (lipoaspirate or solid tissue) and wash it thoroughly with PBS 3-5 times to remove blood contaminants and free lipids [12] [16].
- Enzymatic Digestion: Incubate the washed tissue with a pre-warmed collagenase solution (e.g., 0.1% Liberase in PBS) under controlled agitation for 1 to 4 hours at 37°C. The optimal condition for bovine AT was 0.1% Liberase for 3 hours [8].
- Digestion Neutralization: Add an equal volume of complete growth medium containing serum to neutralize the enzyme activity [12] [16].
- Centrifugation and SVF Pellet Collection: Centrifuge the digested mixture at 800 x g for 10 minutes. This will yield a pellet (the SVF) topped by a layer of adipocytes and lipids. Carefully aspirate the floating adipocytes, lipids, and supernatant [12] [16].
- Red Blood Cell Lysis (Optional): Resuspend the SVF pellet in 160mM NH₄Cl and incubate for 5-10 minutes at room temperature to lyse red blood cells. Centrifuge again at 400 x g for 10 minutes [16].
- Filtering and Plating: Resuspend the final pellet in PBS, filter the cell suspension through a 100µm nylon mesh, and then through a 40µm mesh. Centrifuge, resuspend in growth medium, and plate the cells [12] [16].
The optimization of enzymatic digestion is critical for yield, as illustrated in the following comparative workflow:
Perinatal Tissues: Umbilical Cord Wharton's Jelly Explant and Enzymatic Protocol
The umbilical cord, particularly Wharton's Jelly, is a rich source of primitive MSCs. Two primary methods are employed.
Materials & Reagents:
- Fresh human umbilical cord
- Hypochlorite solution or other disinfectants [16]
- DPBS without Ca²⁺/Mg²⁺
- Collagenase Type I or IV [3] [16]
- Trypsin-EDTA (for enzymatic method)
- Explant growth medium (DMEM with 10-20% FBS)
Procedure: A. Explant Culture Method (Enzyme-Free) [15]
- Cord Preparation: Wash the umbilical cord thoroughly in a hypochlorite solution, followed by multiple rinses in PBS to disinfect the surface and remove blood [16] [15].
- Dissection and Explant Placement: Dissect the cord to expose the Wharton's Jelly. Minced the Wharton's Jelly into small fragments (1-2 mm³).
- Tissue Plating: Place the explants directly onto a culture dish, allowing them to adhere for a short period. Gently add a small volume of complete growth medium to avoid dislodging the explants.
- Culture and Outgrowth: Incubate the explants for 5-7 days. Migratory, fibroblast-like MSCs will grow out from the explants. The medium can be refreshed after this period.
- Explant Removal and Expansion: Once a sufficient outgrowth of cells is observed, remove the explants and continue to culture and expand the adherent MSCs.
B. Enzymatic Digestion Method [3] [16]
- Cord Preparation and Dissection: Follow the same initial washing and dissection steps as in the explant method.
- Enzymatic Digestion: Mince the Wharton's Jelly tissue finely and incubate with a collagenase solution (e.g., 0.1% collagenase) for several hours at 37°C on a shaker.
- Digestion Neutralization and Filtering: Neutralize the enzyme with serum-containing medium. Filter the resulting cell suspension through a cell strainer (e.g., 100µm) to remove undigested tissue fragments.
- Cell Seeding and Culture: Centrifuge the filtrate, resuspend the cell pellet in growth medium, and seed into culture flasks.
Table 2: Key Research Reagent Solutions for MSC Isolation
| Reagent Category | Specific Examples | Function in Protocol |
|---|---|---|
| Digestive Enzymes | Collagenase Type I, Liberase, Trypsin-EDTA [12] [8] | Breaks down the extracellular matrix and dissociates tissues to release individual cells, including MSCs. |
| Gradient Media | Ficoll-Paque, Percoll [10] [16] | Separates mononuclear cells (including MSCs) from other cell types based on density during centrifugation. |
| Culture Media | α-MEM, DMEM, DMEM/F12 [11] [15] | Provides nutrients and environment for the selective attachment and expansion of plastic-adherent MSCs. |
| Serum Supplements | Fetal Bovine Serum (FBS), MSC-qualified FBS [11] [15] | Provides essential growth factors, hormones, and adhesion factors that support MSC attachment, proliferation, and viability. |
| Cell Dissociation Agents | Trypsin-EDTA, TrypLE Express [11] [15] | Detaches adherent MSCs from the culture vessel surface for subculturing and downstream applications. |
Post-Isolation Characterization
Following isolation, MSCs must be characterized based on the criteria established by the International Society for Cell & Gene Therapy (ISCT). These include:
- Plastic Adherence: Under standard culture conditions [10] [3].
- Surface Marker Expression: Positive for CD73, CD90, and CD105; negative for CD34, CD45, HLA-DR, and other hematopoietic markers, as confirmed by flow cytometry [10] [12] [17].
- Multilineage Differentiation Potential: Demonstrated ability to differentiate into osteoblasts, adipocytes, and chondrocytes in vitro [11] [17]. Staining with Alizarin Red (osteogenesis), Oil Red O (adipogenesis), and Alcian Blue (chondrogenesis) confirms this potential.
Bone marrow, adipose tissue, and perinatal tissues each provide distinct advantages as sources of MSCs. Bone marrow remains the historical benchmark, adipose tissue offers superior yield and accessibility, and perinatal tissues provide a potent, non-controversial source. The choice of isolation protocol—whether enzymatic digestion or explant culture—directly impacts cell yield, viability, and functional properties. The protocols and data summarized herein provide a foundational framework for researchers to isolate and characterize MSCs from these key sources reliably, ensuring the generation of high-quality cells for regenerative medicine and therapeutic development.
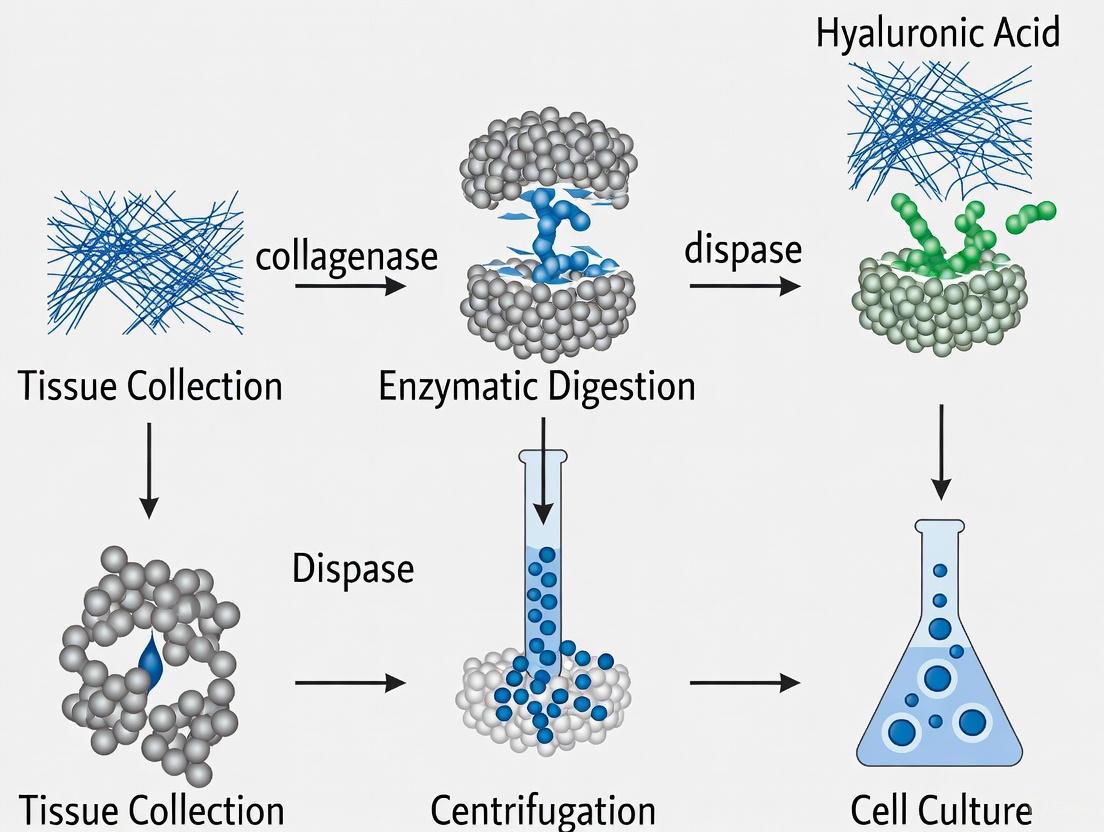
The isolation of Mesenchymal Stromal Cells (MSCs) from tissues is a foundational procedure in regenerative medicine and drug development research. The efficiency of this isolation is paramount, as it directly impacts cell yield, viability, and subsequent functionality. Central to this process is enzymatic digestion, a method that uses specific proteases to disrupt the Extracellular Matrix (ECM)—the complex scaffold of proteins and carbohydrates that encapsulates cells within tissues. The ECM provides structural support and dynamic signaling cues, governing cellular behavior and tissue integrity [18]. Understanding the core principles of how enzymes target and break down key ECM components is essential for optimizing MSC isolation protocols, ensuring the generation of high-quality cell products for therapeutic applications [3] [8].
The Extracellular Matrix: A Proteolytic Landscape
The ECM is not an inert scaffold but a dynamic network of structural proteins and polysaccharides. Its major constituents include fibrillar collagens (providing tensile strength), elastin (conferring elasticity), glycoproteins (like fibronectin and laminin, which facilitate cell adhesion), and proteoglycans (which form hydrated gels) [18]. For cells to be liberated from this network, specific enzymatic activities are required to cleave these components. The process of irreversible proteolysis profoundly impacts development, homeostasis, and disease, and is critically regulated by several families of ECM-degrading proteases [18].
The diagram below illustrates the core mechanism of enzymatic digestion for MSC isolation.
Major Enzyme Families and Their ECM Substrates
ECM-degrading proteases exhibit both functional redundancy and cell-specific specialization, shaped by transcriptional programs and microenvironmental signals [18]. The table below summarizes the key enzyme families used in MSC isolation, their primary substrates, and functional roles.
Table 1: Key Enzyme Families in ECM Digestion for Cell Isolation
| Enzyme Family | Primary ECM Substrates | Mechanism of Action & Role in MSC Isolation |
|---|---|---|
| Collagenases(e.g., Collagenase I, IV) | Fibrillar Collagens (Types I, II, III, IV) | Cleave the triple-helical domain of native collagen, the primary structural protein in many tissues. This is often the rate-limiting step in disrupting the ECM [8] [18]. |
| Serine Proteases(e.g., Trypsin) | Glycoproteins (e.g., Fibronectin, Laminin) | Target peptide bonds and are effective at digesting non-collagenous adhesion proteins. Often used in combination with collagenases to enhance tissue dissociation [3] [8]. |
| Metalloproteinases(e.g., Liberase) | Broad-spectrum: Collagens, Gelatin, Proteoglycans | Enzyme blends (e.g., purified collagenase and neutral protease) designed for high activity and purity, leading to efficient tissue dissociation with potentially better cell viability and yield [8]. |
| Other Proteases(e.g., Dispase, Hyaluronidase) | Specific components (e.g., Laminin, Hyaluronic Acid) | Target specific ECM linkages; often used as supplementary enzymes to create a more complete digestion cocktail [3]. |
Quantitative Evaluation of Enzymatic Protocols
The choice of enzyme, its concentration, and incubation time are critical variables that directly impact the success of MSC isolation. A systematic study evaluating 32 different conditions for isolating bovine adipose tissue-derived MSCs provides valuable quantitative data for protocol optimization [8].
Table 2: Evaluation of Enzymatic Conditions for Bovine Adipose Tissue MSC Isolation
| Enzyme / Mixture | Concentration | Incubation Time | Average Cell Yield (×10^6 cells/g tissue) | Key Findings |
|---|---|---|---|---|
| Collagenase Type I (Coll IA) | 0.1% | 6 hours | ~35 - 130 | A frequently used, reliable method yielding satisfactory results [8]. |
| Collagenase Type I + Trypsin | 0.1% | 3 hours | Not specified | Can yield higher expression of certain stem cell markers in some tissue sources [8]. |
| Liberase (LibTM) | 0.1% | 3 hours | ~30 - 67 | Highest cell yield and low population doubling time; recommended for maximizing yield [8]. |
| Collagenase Type IV | 0.04% | 6 hours / ON / 24 h | No plastic-adherent cells | Certain conditions with this enzyme failed to produce viable, adherent cells [8]. |
The study concluded that for maximizing cell yield when considering MSCs from bovine adipose tissue as a cell source, it is recommended to use 0.1% Liberase for 3 hours [8]. Furthermore, the isolation method can affect not only cell yield but also viability, proliferation potential, and differentiation capacity [8].
Detailed Experimental Protocol: Enzymatic Digestion of Adipose Tissue for MSC Isolation
The following protocol is adapted from optimized conditions evaluated in recent research [8].
Materials and Reagents
Table 3: The Scientist's Toolkit - Essential Reagents for Enzymatic Digestion
| Research Reagent Solution | Function in the Protocol |
|---|---|
| Liberase | The core metalloproteinase blend responsible for digesting collagen and other ECM components. |
| Phosphate Buffered Saline (PBS) | A balanced salt solution used for washing tissue and preparing enzyme solutions. |
| Dulbecco's Modified Eagle Medium (DMEM) | A standard cell culture medium used as a base for the digestion mixture and subsequent cell culture. |
| Fetal Bovine Serum (FBS) | Added to the digestion mixture after incubation to neutralize enzyme activity. |
| Cell Strainer (70-100 µm) | A mesh filter used to remove undigested tissue fragments and obtain a single-cell suspension. |
| Centrifuge Tubes | Tubes used for conducting the digestion process and subsequent centrifugation steps. |
Step-by-Step Methodology
- Tissue Preparation: Aseptically collect adipose tissue (e.g., subcutaneous fat). Wash the tissue thoroughly with PBS containing antibiotics (e.g., 1% Penicillin-Streptomycin) to remove blood and contaminants. Mince the tissue into fine pieces (< 1-2 mm³) using sterile scalpels or scissors.
- Enzyme Solution Preparation: Prepare the digestion solution by dissolving Liberase in a serum-free basal medium like DMEM to a final concentration of 0.1% (w/v). Filter-sterilize the solution using a 0.22 µm filter.
- Digestion Process: Transfer the minced tissue into a centrifuge tube and add the prepared 0.1% Liberase solution. The recommended volume is typically 3-5 times the volume of the tissue. Incubate the tube in a water bath or shaking incubator at 37°C for 3 hours with continuous agitation.
- Reaction Neutralization: After incubation, neutralize the enzymatic reaction by adding an equal volume of complete culture medium (e.g., DMEM supplemented with 10% FBS). This step is crucial to prevent over-digestion, which can damage cell surface markers and reduce viability.
- Cell Separation and Washing: Pass the neutralized cell suspension through a 70-100 µm cell strainer to remove undigested tissue debris. Collect the filtrate and centrifuge at 300-400 × g for 10 minutes. Carefully aspirate the supernatant, resuspend the cell pellet in fresh complete medium, and repeat the centrifugation wash step.
- Cell Seeding and Culture: Resuspend the final cell pellet in an appropriate volume of complete culture medium. Seed the cells into culture flasks and incubate at 37°C in a humidified atmosphere with 5% CO₂. Monitor the cultures daily for fibroblast-like, plastic-adherent cells, which should appear within 2-4 days [8]. Passage the cells when they reach 70-80% confluence, typically observing >5 colony-forming units within 4-7 days post-isolation [8].
Critical Considerations for Protocol Optimization
- Tissue Source Specificity: The optimal digestion protocol can vary significantly depending on the tissue source. For instance, while adipose tissue is readily digested, umbilical cord Wharton's jelly or dental pulp may require different enzyme combinations or incubation times [3]. The density and composition of the ECM are key determining factors.
- Functional Validation: Successful isolation must be followed by MSC characterization. This includes demonstrating adherence to plastic, tri-lineage differentiation potential (osteogenic, adipogenic, chondrogenic), and expression of specific surface markers (e.g., CD73, CD90, CD105) while lacking hematopoietic markers, as per International Society for Cell & Gene Therapy (ISCT) guidelines [3].
- Balancing Yield and Viability: While maximizing cell yield is often a goal, particularly for cultured meat production or large-scale therapeutic applications [8], this must be balanced against cell viability and function. Over-digestion can compromise cell integrity, while under-digestion results in low yield.
The enzymatic digestion of the extracellular matrix is a critical, precision-driven step in the isolation of functional MSCs. A deep understanding of the core principles—including the composition of the ECM, the specific actions of different protease families, and the quantitative impact of protocol variables—empowers researchers to tailor their isolation strategies effectively. By applying optimized, validated protocols such as the use of Liberase for adipose tissue, scientists can ensure high cell yield and quality, thereby advancing the reliability and success of downstream applications in regenerative medicine and drug development.
Advantages and Limitations of Enzymatic Digestion Versus Alternative Methods
The isolation of Mesenchymal Stromal Cells (MSCs) is a foundational step in regenerative medicine and translational research. The choice of isolation method profoundly impacts cell yield, viability, phenotypic characteristics, and subsequent functionality. Within this context, enzymatic digestion and mechanical dissociation represent the two primary methodological approaches. This application note provides a detailed comparative analysis of these techniques, framing them within the specific requirements of establishing a robust MSC isolation protocol. It summarizes quantitative data, provides detailed experimental methodologies, and offers visual workflows to guide researchers in selecting and optimizing the optimal isolation strategy for their scientific and therapeutic objectives.
Comparative Analysis: Enzymatic Digestion vs. Mechanical Dissociation
The decision between enzymatic and mechanical isolation is multifactorial, depending on the tissue source, desired cell population, and intended downstream application. The table below provides a systematic comparison of the two core methodologies.
Table 1: Comprehensive Comparison of MSC Isolation Techniques
| Parameter | Enzymatic Digestion | Mechanical Dissociation |
|---|---|---|
| Core Principle | Uses proteolytic enzymes (e.g., Collagenase, Liberase) to degrade the extracellular matrix and release cells. [12] [3] | Relies on physical force (mincing, chopping, agitation) to dissociate tissue and liberate cells. [19] |
| Key Advantages | - Higher Cell Yield: Generates a more homogenous single-cell suspension, maximizing initial cell harvest. [8]- Reproducibility: Offers greater controllability, supporting standardized, large-scale operations. [19]- Efficiency: Faster sample processing with quicker initial cell harvest compared to explant methods. [8] | - Preserves Microenvironment: Better maintains native cell-cell interactions and the tumor/stromal niche. [19]- No Enzyme Cost/Interference: Avoids potential cell surface receptor damage and eliminates reagent costs. [19] |
| Key Limitations | - Cost: High-quality enzymes are expensive. [8]- Potential Cell Damage: Risk of proteolytic damage to cell surface markers, affecting viability and function. [19]- Complexity: Requires optimization of concentration, time, and neutralization. [8] | - Lower Yield & Viability: Can result in higher cell death and a lower initial yield of viable cells. [19]- Heterogeneous Population: Produces a mix of single cells and tissue fragments, which can be less pure. [19] |
| Ideal Application | - Large-scale drug screening requiring high cell numbers and reproducibility. [19]- Protocols where a homogeneous cell population is critical. | - Research focusing on preserving the tumor microenvironment (e.g., patient-derived organoids for personalized medicine). [19]- Studies where enzymatic cost or potential interference is a primary concern. |
Detailed Experimental Protocols
Optimized Enzymatic Digestion for Adipose-Derived MSCs
This protocol is adapted from established methodologies for isolating MSCs from solid adipose tissue and lipoaspirate. [12] [8] A systematic evaluation of 32 conditions identified the use of Liberase at 0.1% for 3 hours as optimal for bovine adipose tissue, providing the highest cell yield in combination with low population doubling time. [8]
Materials & Reagents:
- Phosphate-Buffered Saline (PBS), sterile
- Liberase (or alternative such as Collagenase Type I)
- Complete Growth Medium (e.g., DMEM/F12 supplemented with Fetal Bovine Serum (FBS) and antibiotics)
- Centrifuge Tubes
- Cell Strainer (70-100 µm)
Step-by-Step Procedure:
- Wash: Transfer the adipose tissue sample to a sterile container. Wash thoroughly with PBS to remove blood contaminants and debris. [12]
- Mince: Using sterile instruments, mince the washed tissue into fine pieces (< 1-2 mm³) to increase the surface area for enzyme action.
- Digest: Add the 0.1% Liberase solution in PBS to the minced tissue. Use a volume sufficient to submerge the tissue completely.
- Incubate: Place the tube in a shaking incubator or on an orbital shaker within a standard CO₂ incubator. Incubate at 37°C for 3 hours with constant agitation. [8]
- Neutralize: After digestion, add an equal volume of complete growth medium (containing serum) to neutralize the enzyme activity.
- Centrifuge: Centrifuge the cell suspension at 300-400 × g for 10 minutes to pellet the cells.
- Filter & Resuspend: Carefully aspirate the supernatant, including the floating adipocytes and oil. Resuspend the cell pellet (the Stromal Vascular Fraction, SVF) in fresh complete medium and pass it through a 100 µm cell strainer to remove residual tissue fragments. [12]
- Seed & Culture: Plate the filtered cell suspension in culture flasks and incubate at 37°C with 5% CO₂. AD-MSCs are selected by their adherence to plastic, and non-adherent cells can be removed during the first medium change after 24-48 hours. [12]
Mechanical Dissociation Workflow
This method serves as an alternative for tissues where preserving the native microenvironment is a priority. [19]
Materials & Reagents:
- Phosphate-Buffered Saline (PBS), sterile
- Complete Growth Medium
- Scalpels or Surgical Blades
- Mechanical Disruptor (e.g., GentleMACS Dissociator) or mortar and pestle
Step-by-Step Procedure:
- Wash and Mince: Wash the tissue sample extensively with PBS. Using sterile scalpels, mince the tissue into a fine pulp.
- Further Disruption: Transfer the minced tissue to a tube containing PBS or growth medium.
- Option A (Manual): Use a pipette to repeatedly triturate the mixture vigorously. Alternatively, use a sterile mortar and pestle for further grinding.
- Option B (Automated): Transfer the minced tissue to a C-tube and process using a mechanical dissociator according to the manufacturer's program for soft tissues.
- Settle and Collect: Allow the suspension to settle briefly so that large fragments sediment. Collect the supernatant containing the released cells and tissue clusters.
- Centrifuge and Seed: Centrifuge the supernatant at a low speed (200-300 × g) for 5-10 minutes. Resuspend the pellet in complete growth medium and seed into culture flasks.
Workflow and Decision-Making Visualization
The following diagram illustrates the logical decision-making process for selecting an appropriate MSC isolation method based on core research objectives.
The Scientist's Toolkit: Essential Research Reagents
Successful isolation and characterization of MSCs rely on a defined set of reagents and tools. The following table details the key materials required for the protocols described in this note.
Table 2: Essential Reagents for MSC Isolation and Characterization
| Reagent/Material | Function/Application | Examples & Notes |
|---|---|---|
| Collagenase Type I / Liberase | Proteolytic enzyme blend for digesting collagen in the extracellular matrix during enzymatic isolation. [12] [8] | Liberase is a purified, GMP-grade enzyme mixture often yielding higher purity and efficiency. [8] |
| Trypsin-EDTA | Proteolytic enzyme used for routine subculturing and passaging of adherent MSCs post-isolation. | Not typically used for primary tissue dissociation due to higher cytotoxicity. |
| Density Gradient Medium | Separation of mononuclear cells (like MSCs) from other cellular components based on density. | Ficoll-Paque; Percoll. Often used for bone marrow but less so for adipose tissue. |
| Fetal Bovine Serum (FBS) | Critical supplement in growth media to support MSC adhesion, proliferation, and viability. | Serum-free alternatives are available but require validation for primary isolation. |
| Flow Cytometry Antibodies | Immunophenotyping to confirm MSC identity per ISCT criteria (positive & negative markers). [3] | CD73, CD90, CD105 (Positive); CD34, CD45, HLA-DR (Negative). |
| Tri-lineage Differentiation Kits | Functional validation of MSC multipotency (adirogenic, osteogenic, chondrogenic). [8] | Commercially available kits include specific induction and staining media. |
| Cell Strainers | Removal of undigested tissue fragments and debris to obtain a single-cell suspension. | Typically 70 µm and 100 µm mesh sizes used sequentially or individually. |
The isolation of viable mesenchymal stromal cells (MSCs) from native tissues represents a critical initial step in regenerative medicine, drug development, and basic biological research. Enzymatic dissociation enables researchers to break down the extracellular matrix (ECM) that encases cells within tissues, facilitating the release of functional, viable cells for downstream applications. The selection of appropriate enzymes directly influences cell yield, viability, proliferation potential, and the preservation of critical surface markers. This article provides a comprehensive overview of three key enzymatic tools—collagenase, trypsin, and Liberase—within the specific context of MSC isolation research. We detail their mechanisms, applications, and provide optimized protocols to guide researchers in selecting the most effective dissociation strategy for their experimental needs.
The efficacy of MSC isolation protocols hinges on understanding the composition of the target tissue. Adipose tissue, bone marrow, and other MSC-rich sources contain substantial amounts of collagen, the primary structural protein in the ECM. Collagen molecules form a unique triple-helical structure that is resistant to most proteases. Only specific enzymes, termed collagenases, can initiate the degradation of native, fibrillar collagen under physiological conditions, making them indispensable for tissue dissociation [20]. The subsequent breakdown of other ECM components and cell-adhesion proteins is then facilitated by broader-spectrum proteases.
Enzyme Fundamentals: Mechanisms and Targets
Collagenase
- Source and Classification: Commercial collagenase used in cell isolation is predominantly sourced from the bacterium Clostridium histolyticum [20] [21]. These enzymes are metalloproteases (zinc-dependent) and belong to the M9 family of peptidases. C. histolyticum produces two main classes of collagenases, Class I (product of the colG gene) and Class II (product of the colH gene), which have distinct but complementary substrate preferences [20] [22].
- Mechanism of Action: Collagenases are uniquely capable of cleaving the triple-helical domains of native collagen. They specifically recognize and hydrolyze peptide bonds within the sequence Pro-X-Gly-Pro, where X is often a neutral amino acid, cutting between X and Gly [20]. This initial cleavage unravels the robust collagen structure, allowing other proteases to further degrade the ECM.
- Commercial Preparations: It is crucial to understand that commercially available "collagenase" is not a pure substance but a complex mixture containing both Class I and II collagenases along with other proteases such as clostripain, a neutral protease, and a trypsin-like enzyme [20] [22]. These supplemental proteolytic activities synergistically digest non-collagenous proteins in the tissue. Products are thus formulated into different "types" (I, II, III, IV, V, etc.) with balanced or enriched activities tailored for specific tissues [20].
Trypsin
- Source and Classification: Trypsin is a serine protease derived from pancreatic sources. Its enzymatic activity relies on a catalytic triad of histidine, aspartate, and serine [23].
- Mechanism of Action: Trypsin exhibits high specificity for cleaving peptide bonds at the carboxyl side of the basic amino acids lysine (Lys) and arginine (Arg) [23]. It functions as an endopeptidase, cutting within polypeptide chains rather than at the terminals.
- Role in Tissue Dissociation: In tissue dissociation protocols, trypsin primarily targets intercellular proteins, such as fibronectin and laminin, that facilitate cell-cell and cell-ECM adhesion [23]. However, its aggressive proteolytic nature can damage cell surface receptors and antigens, potentially affecting downstream cell functionality and viability [23]. It is often used in combination with other enzymes like collagenase or as a component of proprietary blends.
Liberase
- Composition: Liberase is a proprietary, highly purified enzyme blend designed for consistent and gentle tissue dissociation. While its exact formulation is not fully disclosed, it is known to consist of highly purified collagenase I and II, combined with a neutral protease such as dispase or thermolysin [8] [21] [24].
- Mechanism of Action: Liberase enzymes work synergistically to degrade both collagenous and non-collagenous components of the ECM. The collagenases disrupt the collagen backbone, while the neutral protease cleaves other proteins without the trypsin-like specificity for lysine and arginine, which is considered a gentler alternative [21].
- Key Advantages: The primary benefits of Liberase are its high purity, which reduces lot-to-lot variability, and its gentler action on cell membranes, leading to improved cell viability and better preservation of sensitive surface markers [8] [24].
The following diagram illustrates the primary mechanisms of action for each enzyme and their synergistic relationship in a blended protocol:
Comparative Analysis of Enzyme Performance in MSC Isolation
The choice of enzyme and digestion protocol has a direct and significant impact on the success of MSC isolation. A systematic evaluation of different conditions is essential for optimization.
Table 1: Comparative Evaluation of Enzymes for Bovine Adipose-Derived MSC Isolation [8]
| Enzyme / Blend | Concentration | Incubation Time | Average Cell Yield (×10⁶ cells/g tissue) | Key Observations |
|---|---|---|---|---|
| Liberase | 0.1% | 3 hours | 30.5 - 67.1 | Highest cell yield; low population doubling time; successful isolation from most donors. |
| Collagenase Type I | 0.1% | 3 hours | Baseline (for comparison) | Lower cell yield compared to Liberase under identical conditions. |
| Collagenase Type I + Trypsin | 0.1% | 3 hours | Varied | Commonly used combination; yield highly dependent on specific tissue type. |
| Collagenase Type IV | 0.04% | 6 hours / ON / 24 hours | 0 (No adherent cells) | Unsuitable for bovine adipose tissue under these conditions. |
Interpretation of Experimental Data
A 2024 study systematically evaluated 32 isolation conditions for bovine adipose-derived MSCs, providing critical quantitative data for protocol selection [8]. The findings demonstrated that Liberase at a concentration of 0.1% with a 3-hour incubation time yielded the highest cell yield while maintaining low population doubling times, indicating robust cell health and proliferation potential. Notably, varying enzyme concentration and incubation time did not always result in statistically significant differences in yield, but a trend was observed where higher concentrations (0.1%) combined with shorter incubation times (3-6 hours) generally produced superior results [8]. This highlights the importance of avoiding over-digestion, which can compromise cell viability.
Application Notes and Protocols
Optimized Protocol for MSC Isolation from Adipose Tissue
Based on recent comparative studies, the following protocol is recommended for the isolation of MSCs from bovine subcutaneous adipose tissue, with principles applicable to other sources [8].
Objective: To efficiently isolate viable mesenchymal stromal cells from adipose tissue with high yield and purity for downstream culture and differentiation experiments.
Materials:
- Research Reagent Solutions:
- Liberase: A purified blend of collagenase I and II and a neutral protease (e.g., dispase or thermolysin) [8] [21].
- Collagenase Type I: A balanced mixture of collagenase classes and secondary proteases, suitable for epithelial tissue, liver, lung, and adipose tissue [20] [22].
- Collagenase Type II: Formulation with higher clostripain (protease) activity, ideal for tough tissues like bone, heart, and cartilage [20] [22].
- Trypsin-EDTA: A solution of trypsin and the chelator EDTA, used to dissociate adherent cells from culture surfaces by targeting cell-adhesion proteins [23].
- Phosphate Buffered Saline (PBS): A balanced salt solution for washing and diluting reagents.
- Cell Culture Medium: Complete medium (e.g., DMEM/F12 supplemented with fetal bovine serum (FBS) and antibiotics) to stop enzymatic activity and sustain cells.
Procedure:
- Tissue Preparation: Aseptically collect subcutaneous adipose tissue. Wash thoroughly with PBS to remove blood clots and debris. Mince the tissue into fine pieces (approximately 1-2 mm³) using sterile scalpels.
- Enzymatic Digestion:
- Prepare the digestion solution by dissolving Liberase in a balanced salt solution or PBS to a final concentration of 0.1% (w/v) [8].
- Transfer the minced tissue to the enzyme solution, using a ratio of 1g of tissue per 3-5 mL of enzyme solution.
- Incubate the mixture for 3 hours at 37°C with constant agitation (e.g., on a shaker or orbital mixer) [8].
- Reaction Termination and Cell Harvest:
- After incubation, neutralize the enzyme by adding an equal volume of complete cell culture medium containing serum (e.g., 10% FBS).
- Pass the cell suspension through a sterile cell strainer (e.g., 70-100 µm) to remove undigested tissue fragments and debris.
- Centrifuge the filtered suspension at 300-400 × g for 10 minutes to pellet the cells.
- Cell Seeding and Culture:
- Aspirate the supernatant and resuspend the cell pellet in fresh complete culture medium.
- Seed the cells into culture flasks at the desired density.
- Place the flasks in a 37°C, 5% CO₂ incubator and allow the cells to adhere.
- Protocol Validation: Replace the medium after 24-48 hours to remove non-adherent cells. Monitor for the emergence of colony-forming unit fibroblasts (CFU-Fs), which indicate successful MSC isolation. Passage cells when they reach 70-80% confluence.
Troubleshooting Notes:
- Low Cell Yield: Ensure the enzyme is fully dissolved and active. Consider increasing the enzyme concentration slightly or extending the incubation time, but monitor cell viability closely.
- Poor Cell Viability: Reduce the enzyme concentration or incubation time. The presence of serum in the neutralization medium is critical to rapidly halt proteolytic activity.
- Calcium Dependence: Collagenase activity is calcium-dependent. Avoid using calcium-chelating agents like EDTA or EGTA in the digestion buffer, as they will inhibit the enzyme [21].
The workflow for this optimized protocol is summarized below:
Guide to Collagenase Type Selection
Given that "collagenase" products are complex mixtures, selecting the appropriate type is crucial for isolating MSCs from different tissues.
Table 2: Collagenase Type Selection Guide for MSC Isolation [20] [22]
| Collagenase Type | Enzymatic Component Characteristics | Recommended Tissue Applications for MSC Isolation |
|---|---|---|
| Type I | Balanced activity of collagenase, clostripain, and trypsin-like enzymes. | Adipose tissue, adrenal tissue, liver, lung, epithelial tissue. |
| Type II | Higher clostripain (neutral protease) activity. | Bone, heart, liver, thymus, salivary gland, cartilage. |
| Type III | Lower hydrolytic activity of secondary proteases. | Mammary tissue (e.g., breast tissue). |
| Type IV | Low trypsin-like activity. | Pancreatic islets (Note: unsuitable for bovine adipose tissue [8]). |
| Type V | Higher collagenase and caseinase activities, lower tryptic activity. | Suitable for islet preparation. |
The strategic selection and application of dissociation enzymes are fundamental to successful MSC research. While traditional reagents like collagenase and trypsin remain widely used, modern, highly purified blends like Liberase offer demonstrable advantages in terms of cell yield, viability, and protocol consistency, as evidenced by recent comparative studies [8].
The "ideal" protocol is context-dependent. Researchers must consider the tissue of origin, the specific MSC subpopulation targeted, and the intended downstream application. For instance, while a Collagenase Type I/Trypsin blend might be effective for some tissues, the data strongly supports the use of Liberase at 0.1% for 3 hours for the isolation of MSCs from bovine adipose tissue [8]. For denser tissues like bone, a collagenase with higher supplemental protease activity (e.g., Type II) may be more appropriate [20] [22].
Future directions in enzymatic dissociation will likely focus on the development of even more specific and gentle recombinant enzyme blends, further reducing batch-to-batch variability and enhancing the recovery of functionally pristine cells. As the field of regenerative medicine and cultured meat production advances, the optimization of these critical first steps in cell isolation will remain a cornerstone of successful research and development.
Step-by-Step: Tissue-Specific Enzymatic Digestion Protocols
Mesenchymal Stromal Cells (MSCs) are multipotent cells with significant potential in regenerative medicine and drug development due to their immunomodulatory properties, differentiation capacity, and relative ease of isolation from various tissues [3]. The transition from raw tissue to a stable, characterized primary culture is a critical foundational step in MSC research. This application note details a standardized workflow for the isolation and initial culture of MSCs from adipose tissue and umbilical cord, two of the most common and abundant sources [3] [16]. The protocols are framed within a broader thesis on optimizing enzymatic digestion for MSC isolation, providing researchers with validated methodologies to ensure cell yield, viability, and phenotypic fidelity.
MSCs can be isolated from a wide range of adult and perinatal tissues. The choice of source tissue can influence the yield, proliferation rate, and secretome of the resulting primary cells [25].
- Adipose Tissue: Typically obtained from lipoaspirate or surgical waste, adipose tissue is a rich source of MSCs (often termed Adipose-Derived Stromal Cells or ADSCs). Its primary advantages include high abundance and a high frequency of MSCs per gram of tissue [8] [25].
- Umbilical Cord: Perinatal tissues like the umbilical cord (UC) are medical waste products, posing fewer ethical concerns. The Wharton's Jelly within the UC is a robust source of MSCs (WJ-MSCs) known for high proliferative capacity [3].
Pre-processing Steps:
- Adipose Tissue: The lipoaspirate is washed multiple times with Phosphate Buffered Saline (PBS) to remove blood, saline, and lysed adipocytes. The upper oil layer and lower liquid layer are aspirated and discarded, leaving the intact adipose tissue for downstream processing [16].
- Umbilical Cord: The cord is washed thoroughly in a hypochlorite solution or PBS to decontaminate, then stored briefly in a medium containing antibiotics and serum before further dissection and digestion [16].
Isolation Techniques: Explant vs. Enzymatic Digestion
Two primary methods are employed to isolate MSCs from tissue: the explant method and enzymatic digestion. The choice between them involves a trade-off between cell yield, processing time, and potential impact on cell characteristics [25].
The following workflow outlines the decision points and procedures for these two fundamental isolation pathways:
Explant Method
This technique relies on the innate migratory capacity of MSCs. Fragments of tissue are anchored to a culture surface, allowing cells to migrate out spontaneously.
- Workflow: The middle layer of washed adipose tissue or fragments of Wharton's Jelly are placed in culture dishes with a growth medium supplemented with 10-20% Fetal Bovine Serum (FBS). The plates are left undisturbed for 2-4 weeks, during which MSCs migrate out of the tissue fragments, adhere to the plastic surface, and begin to proliferate [25].
- Advantages and Disadvantages: The explant method is mechanically gentle and avoids the cost and potential cell damage of enzymes. Its main drawback is the significantly longer time required to obtain a sufficient number of cells for primary culture (approximately 10 days to first passage) and a lower overall cell yield compared to enzymatic digestion [8] [25].
Enzymatic Digestion
This method uses enzymes to break down the extracellular matrix (ECM) of the tissue, liberating the individual cells, including the MSC-containing stromal vascular fraction (SVF).
- Workflow: The pre-processed tissue is incubated with an enzyme mixture under controlled conditions (temperature, time, agitation). The reaction is neutralized with a serum-containing medium. The cell suspension is then centrifuged to pellet the SVF, which may be further purified through a density gradient (e.g., Percoll or Ficoll) or filtered to remove debris before plating [8] [16].
- Advantages and Disadvantages: Enzymatic digestion offers higher cell yields in a shorter time (cells can be passaged within 4-7 days) and is more reproducible. However, it requires optimization of enzyme type, concentration, and incubation time to balance yield against potential damage to cell surface markers and viability [8] [25].
Optimizing Enzymatic Digestion for MSC Isolation
The efficiency of enzymatic isolation is highly dependent on the specific protocol. A study on bovine adipose tissue evaluated 32 different conditions to maximize cell yield, providing critical data for protocol selection [8].
Table 1: Evaluation of Enzymatic Conditions for Isolating MSCs from Adipose Tissue [8]
| Enzyme Mixture | Concentration | Incubation Time | Average Cell Yield (cells/g tissue) | Key Findings |
|---|---|---|---|---|
| Collagenase Type I (Coll IA) | 0.1% | 6 hours | > 35 x 10⁶ | A frequently used, reliable standard. |
| Collagenase Type I + Trypsin | 0.1% | 3 hours | Not specified | Comparable yield to Collagenase I alone. |
| Liberase (LibTM) | 0.1% | 3 hours | 30.48 - 67.1 x 10⁶ | Significantly higher yield than Collagenase I; fast. |
| Liberase (LibTM) | 0.1% | 6 hours | > 35 x 10⁶ | High yield, but longer incubation. |
| Liberase (LibTM) | 0.1% | Overnight (ON) | > 35 x 10⁶ | High yield, but extended processing. |
| Collagenase Type IV | 0.04% | 6 hours | No adherent cells | Ineffective under these conditions. |
Key Conclusions from Optimization Studies [8]:
- Enzyme Selection: Liberase at a concentration of 0.1% for 3 hours was identified as the most efficient condition for isolating bovine ADSCs, yielding the highest number of cells with a low population doubling time.
- Concentration and Time: Higher enzyme concentrations (0.1%) combined with shorter incubation times (3-6 hours) generally yielded more cells than lower concentrations or extended incubations.
- Cell Characteristics: Cells isolated under these optimal conditions maintained typical MSC characteristics, including trilineage differentiation potential and expression of standard MSC surface markers.
Detailed Step-by-Step Protocols
This protocol outlines the isolation of the Stromal Vascular Fraction (SVF) from adipose tissue.
- Wash Tissue: Take approximately 250 mL of fat and wash it 3-5 times with an equal volume of PBS. For each wash, agitate and let the tissue separate, then discard the lower aqueous phase until it becomes clear.
- Digest: Add Collagenase solution (e.g., 0.1% Liberase or Collagenase Type I) to the washed tissue. Incubate at 37°C for 1-4 hours on a shaker.
- Neutralize: Add a volume of medium containing 10% FBS to neutralize the enzyme activity.
- Centrifuge: Centrifuge the digested mixture at 800 x g for 10 minutes. This will separate the contents into a pellet (the SVF), a layer of floating adipocytes and lipids, and a liquid supernatant.
- Collect Pellet: Carefully aspirate and discard the floating adipocytes, lipids, and liquid supernatant, leaving the SVF pellet.
- Lyse Erythrocytes: Resuspend the SVF pellet in 160mM NH₄Cl solution and incubate for 10 minutes at room temperature to lyse any remaining red blood cells.
- Centrifuge: Centrifuge at 400 x g for 10 minutes.
- Purify (Optional): Layer the cell suspension on a Percoll or Histopaque density gradient. Centrifuge at 1000 x g for 30 minutes. Mononuclear cells, including MSCs, will form a buffy coat at the interface.
- Wash and Filter: Collect the interface cells, wash twice with PBS, and filter the cell suspension sequentially through 100µM and 40µM nylon meshes to remove cell clumps and debris.
- Plate Cells: Resuspend the final cell pellet in a growth medium (e.g., DMEM with 40% FBS) and plate in culture flasks. Incubate at 37°C in a 5% CO₂ incubator.
This protocol focuses on isolating MSCs from the Wharton's Jelly of the umbilical cord.
- Decontaminate: Wash the intact umbilical cord in a hypochlorite solution (diluted 1:3), followed by several rinses in PBS.
- Dissect and Store: The cord can be stored for up to 12 hours in a solution like 10% FBS/DMEM-low glucose.
- Digest: Inject 0.1% collagenase in PBS into the vein and arteries of the cord. Incubate the entire cord for 20 minutes at 37°C.
- Harvest Cells: Inject 5mL of DMEM-low glucose with 10% FBS into the cord and massage the tissue vigorously to dissociate and flush out the cells.
- Centrifuge: Collect the cell suspension and centrifuge at 300 x g for 10 minutes.
- Plate Cells: Resuspend the pellet in a culture medium and plate in culture flasks. Incubate at 37°C in a 5% CO₂ incubator.
Establishing Primary Culture
Once isolated, cells are plated to establish the primary culture, termed passage 0 (P0).
- Culture Conditions: Cells are maintained in a specialized MSC growth medium (e.g., PromoCell MSC Growth Medium) or a basal medium like DMEM or αMEM, supplemented with FBS (10-20%), L-glutamine, and antibiotics [25] [26]. Incubation is standardly performed at 37°C in a humidified atmosphere of 5% CO₂.
- Surface Coating: Some defined, serum-free media require culture vessels to be pre-coated with human or bovine fibronectin (e.g., 10 µg/cm²) to facilitate cell adhesion and spreading [26].
- Medium Changes: The medium should be replaced for the first time 3-4 hours after initial plating to remove non-adherent cells and debris, and subsequently every 2-3 days thereafter [26].
- Subcultivation: When cells reach 70-90% confluence, they can be subcultured. It is recommended to use a gentle dissociation reagent like Accutase for detachment. Cells are centrifuged at 220 x g for 3 minutes and reseeded at a recommended density of 1-2.5 x 10³ cells/cm² [25] [26].
The Scientist's Toolkit: Essential Reagents and Materials
Table 2: Key Research Reagent Solutions for MSC Isolation and Culture
| Reagent/Material | Function/Application | Examples / Notes |
|---|---|---|
| Collagenase | Digests collagen in the extracellular matrix to dissociate tissues. | Type I is most common; Type IV also used. |
| Liberase | A purified enzyme blend (Collagenase & Neutral Protease). | Can offer higher yield and viability than traditional collagenase [8]. |
| Trypsin | Protease often used in combination with collagenase. | Can help dissociate cell clusters post-collagenase digestion. |
| Fetal Bovine Serum (FBS) | Supplements growth medium; provides adhesion factors, hormones, and nutrients. | Critical for explant method and neutralizing enzymes. |
| Defined MSC Growth Medium | Serum-free medium for expansion. | Often requires fibronectin coating of flasks (e.g., MSC Growth Medium DXF) [26]. |
| Percoll / Ficoll-Paque | Density gradient medium for purifying mononuclear cells from the stromal vascular fraction. | Used to separate MSCs from red blood cells and debris [16]. |
| Fibronectin | Extracellular matrix protein for coating culture vessels. | Essential for cell adhesion in some serum-free media [26]. |
| Accutase | Enzyme blend for cell detachment. | Gentler than trypsin, recommended for subculturing MSCs [26]. |
| Antibiotics (Pen/Strep) | Prevents bacterial contamination in primary cultures. | Standard supplement in wash and growth media. |
A successful transition from tissue to primary MSC culture hinges on a well-considered and executed workflow. The choice between the explant and enzymatic methods depends on the research priorities of yield, speed, and simplicity. For enzymatic protocols, optimization of parameters such as enzyme type, concentration, and digestion time is paramount for maximizing the isolation of viable, functionally competent MSCs. By adhering to these detailed protocols and utilizing the essential toolkit of reagents, researchers can establish a robust foundation for downstream applications in drug development, cellular characterization, and regenerative medicine.
Within the broader scope of thesis research on enzymatic digestion protocols for isolating mesenchymal stromal cells (MSCs), the optimization of initial digestion parameters is a critical determinant of success. Adipose-derived MSCs (AD-MSCs) offer a promising cell source for regenerative medicine and drug development due to their easy accessibility and multipotent potential [12]. The enzymatic liberation of the stromal vascular fraction (SVF) from adipose tissue is the foundational step, with its efficiency directly impacting downstream experimental outcomes. This application note details a standardized, quantitative approach to evaluate key enzymatic digestion variables—enzyme selection, concentration, and incubation time—to ensure high yields of viable, functional AD-MSCs for research and therapeutic development.
Experimental Protocol: Evaluating Enzymatic Digestion Conditions
Reagents and Materials
- Adipose Tissue: Subcutaneous adipose tissue, either as solid tissue or lipoaspirate, collected aseptically.
- Enzymes:
- Collagenase Type I
- Collagenase Type I + Trypsin
- Liberase
- Collagenase Type IV
- Buffers and Media: Phosphate-Buffered Saline (PBS), sterile; Complete Growth Medium (e.g., α-MEM or DMEM), supplemented with fetal bovine serum (FBS) or human platelet lysate and antibiotics.
- Equipment: Biological safety cabinet, CO₂ incubator, water bath or heater with agitation, centrifuge, sterile filtration units (100 µm, 70 µm), hemocytometer or automated cell counter, tissue culture plasticware.
Step-by-Step Methodology
- Tissue Preparation: Minced adipose tissue is washed extensively with PBS to remove blood contaminants and red blood cells [12].
- Enzymatic Digestion: The washed tissue is subjected to digestion using the selected enzyme or enzyme mixture. The following conditions should be tested in parallel, with digestion performed under controlled temperature (37°C) and constant agitation [12] [8].
- Reaction Neutralization: The enzymatic activity is neutralized by adding an equal volume of complete growth medium containing serum [12].
- Stromal Vascular Fraction (SVF) Isolation: The digested tissue is centrifuged (e.g., 1200 × g for 10 minutes). The resulting pellet, the SVF, is resuspended in growth medium [12].
- Filtration and Seeding: The cell suspension is filtered through a 100 µm sterile filter to remove residual tissue fragments and undigested debris. The filtrate is then passed through a 70 µm filter to obtain a single-cell suspension. The cells are seeded in culture flasks [12].
- Cell Culture and Expansion: Cultures are maintained in a humidified incubator at 37°C with 5% CO₂. The medium is changed after 48-72 hours to remove non-adherent cells, and subsequently every 3-4 days until 70-80% confluence is reached [12].
Workflow Visualization
The diagram below outlines the experimental workflow for isolating and characterizing Adipose-Derived Mesenchymal Stromal Cells (AD-MSCs).
Data Presentation: Comparative Analysis of Enzymatic Conditions
A systematic evaluation of 32 isolation conditions for bovine adipose tissue revealed critical insights into optimizing cell yield and quality. The data below summarizes key findings for the most effective parameters.
Table 1: Impact of Enzyme Type and Concentration on Cell Yield
| Enzyme / Mixture | Concentration | Incubation Time | Average Cell Yield (×10⁶ cells/g tissue) | Key Observations |
|---|---|---|---|---|
| Liberase | 0.1% | 3 hours | 30.5 - 67.1 [8] | Highest cell yield, low population doubling time [8] |
| Collagenase Type I | 0.1% | 6 hours | > 35 [8] | Reliable yield, commonly used in standardized protocols [12] [8] |
| Collagenase Type I + Trypsin | Not Specified | Not Specified | Variable | Can improve yield in some species; requires optimization [8] |
| Collagenase Type IV | 0.04% | 6-24 hours | No plastic-adherent cells | Not recommended for bovine AT-MSC isolation [8] |
Table 2: Effect of Incubation Time on Isolation Success with 0.1% Liberase
| Incubation Time | Success Rate (Isolations from 8 donors) | Days to >5 Colony Forming Units |
|---|---|---|
| 3 hours | 5/8 | 5 days [8] |
| 6 hours | 5/8 | 5 days [8] |
| Overnight (ON) | 5/8 | >5 days [8] |
The Scientist's Toolkit: Key Research Reagent Solutions
Table 3: Essential Reagents for AD-MSC Isolation and Characterization
| Reagent / Material | Function in the Protocol | Key Considerations for Selection |
|---|---|---|
| Type I Collagenase | Enzymatically digests collagen matrix in adipose tissue to release SVF [12] [8]. | Purity and activity vary between suppliers; critical for protocol reproducibility. |
| Liberase | Proprietary enzyme blend (Collagenase + Neutral Protease). Designed for GMP-compliant cell isolation [8]. | Offers high efficiency and lot-to-lot consistency; recommended for high-yield isolation [8]. |
| PBS (without Ca²⁺/Mg²⁺) | Washing buffer to remove blood cells and contaminants from tissue [12]. | Absence of calcium and magnesium prevents cell clumping and premature enzyme activation. |
| Complete Growth Medium | Neutralizes enzymatic activity; supports cell growth and expansion [12] [27]. | Serum-free supplements (e.g., platelet lysate) enhance reproducibility and clinical relevance [27]. |
| Antibodies for Flow Cytometry | Immunophenotyping to confirm MSC identity (positive: CD73, CD90, CD105; negative: CD34, HLA-DR) [12] [28] [17]. | Panels should be validated for the specific species and tissue source. |
Characterization of Isolated Cells
Following isolation, AD-MSCs must be characterized based on the criteria established by the International Society for Cell & Gene Therapy (ISCT). The following diagram illustrates the logical framework for the multi-parameter characterization of Mesenchymal Stromal Cells (MSCs) to confirm their identity.
- Immunophenotyping: Flow cytometry analysis must demonstrate expression of typical MSC surface markers (e.g., CD73, CD90, CD105) and lack of expression of hematopoietic markers (e.g., CD34, HLA-DR) [12] [28] [17].
- Trilineage Differentiation: The multipotency of isolated cells is confirmed by inducing differentiation into adipocytes, osteocytes, and chondrocytes, verified by specific staining (Oil Red O for lipids, Alizarin Red for calcium, and Alcian Blue for proteoglycans, respectively) [8] [17].
- Functional Assays: Additional functional characterization, such as T-cell proliferation suppression assays, can be performed to confirm immunomodulatory capacity, a key therapeutic property of MSCs [27].
This protocol provides a detailed framework for evaluating and optimizing the enzymatic digestion of adipose tissue to isolate high-quality AD-MSCs. Data demonstrates that enzyme selection (Liberase), concentration (0.1%), and incubation time (3 hours) are pivotal parameters for maximizing cell yield while maintaining viability and functionality. Standardizing this initial step is fundamental for ensuring the reproducibility and reliability of all subsequent research within a thesis focused on MSC isolation and application, ultimately contributing to robust and translatable findings in regenerative medicine and drug development.
Mesenchymal stromal cells (MSCs) derived from the Wharton's jelly (WJ) of the umbilical cord represent a promising tool for regenerative medicine and drug development due to their high proliferation capacity, multipotency, and low immunogenicity [29] [30]. Unlike bone marrow-derived MSCs, their collection is non-invasive and raises no ethical concerns, as they are obtained from medical waste following birth [29] [31]. The isolation of high-quality WJ-MSCs is a critical first step for research and clinical applications, primarily achieved through two distinct methodologies: enzymatic digestion and explant culture [28] [32]. This protocol details both approaches, providing a comparative analysis to guide researchers in selecting and optimizing their isolation procedures within the broader context of standardized mesenchymal stromal cell isolation research.
Comparative Analysis of Isolation Methods
The choice between enzymatic digestion and explant culture involves a trade-off between cell yield, processing time, standardization potential, and technical simplicity. The following table summarizes the core characteristics of each method.
Table 1: Key Comparison Between Explant and Enzymatic Digestion Methods for WJ-MSC Isolation
| Feature | Explant Method | Enzymatic Digestion Method |
|---|---|---|
| Basic Principle | Cells migrate out from tissue fragments adhered to a culture surface [29] [31]. | Tissue matrix is broken down by enzymes to release individual cells [29] [30]. |
| Primary Cell Yield | Lower yield per gram of tissue [33] [30]. | Higher and more immediate yield of primary cells [33] [30]. |
| Time to Primary Culture | Longer (7-21 days for cell outgrowth) [33] [31]. | Shorter (cells available for culture immediately post-digestion) [30] [32]. |
| Technical Complexity | Simple, requires minimal reagents [29] [34]. | More complex, requires optimization of enzyme type, concentration, and time [33] [30]. |
| Standardization Potential | Lower, due to reliance on cell migration [32]. | Higher, offers a more robust and reproducible protocol [30] [32]. |
| Risk of Contamination | Reduced risk of biological contamination from enzymes [29]. | Potential risk if enzymes are not sterile or GMP-compliant [30]. |
| Cell Phenotype & Differentiation | No significant differences post-passaging [30] [32]. | No significant differences post-passaging [30] [32]. |
Beyond these core characteristics, quantitative data from direct comparative studies further illuminates the differences in performance. One study found that the explant method had a significantly longer culture cycle and lower yield of primary cells per centimeter of umbilical cord compared to enzymatic methods [33]. Furthermore, subculture adherence occurred faster with the explant method than with conventional enzymatic digestion, though a modified enzymatic protocol eliminated this difference [33].
Table 2: Quantitative Outcomes from Comparative Studies of WJ-MSC Isolation Methods
| Parameter | Explant Method | Conventional Enzymatic Digestion | Modified Enzymatic Digestion |
|---|---|---|---|
| Primary Cell Yield | Lower yield per cm of cord [33] | Higher than explant [33] | Highest yield (e.g., 0.4 PZ U/mL Collagenase NB6 for 3h) [33] [30] |
| Time to Primary Culture | 7-21 days [33] [31] | Several hours digestion [33] | Several hours digestion (e.g., 16-20h for 0.2% Collagenase II) [33] |
| Time to Subculture Adherence | Faster than conventional digestion [33] | Slower than explant [33] | No significant difference from explant [33] |
Detailed Experimental Protocols
Explant Method Protocol
The explant method is valued for its simplicity and minimal requirement for specialized reagents [31] [34].
Materials:
- Phosphate-Buffered Saline (PBS), sterile
- Alpha-Minimal Essential Medium (α-MEM) or Dulbecco's Modified Eagle Medium (DMEM)
- Fetal Bovine Serum (FBS), 10-15%
- Antibiotic-Antimycotic solution (e.g., Penicillin-Streptomycin)
- Tissue culture flasks/dishes
Procedure:
- Sample Pre-processing: Transfer the umbilical cord to a sterile laminar flow hood. Rinse the cord thoroughly with PBS containing 1% antibiotics to remove residual blood [31] [34].
- Vessel Removal and Dissection: Using sterile forceps and scalpels, dissect the cord to expose the Wharton's jelly. Carefully remove the two arteries and one vein. Dice the remaining Wharton's jelly into small fragments of approximately 2-4 mm³ [31].
- Culture Initiation: Transfer the tissue fragments into a culture flask, placing them at small intervals. Add a minimal volume of complete culture medium (e.g., α-MEM supplemented with 10-15% FBS and antibiotics) to just cover the fragments. Do not disturb the flask for 5-7 days to allow the fragments to adhere [31] [34].
- Medium Replacement and Outgrowth: After 5-7 days, carefully add more complete medium without dislodging the explants. Replace half of the medium every 3-4 days thereafter.
- Explant Removal and Passaging: After 10-14 days, or when a substantial outgrowth of fibroblast-like, adherent cells is observed surrounding the explants, carefully remove the tissue pieces. Allow the adherent MSCs to reach 70-80% confluence before passaging them using a standard trypsin-EDTA solution for subsequent expansion and characterization [31].
Enzymatic Digestion Protocol
This method is preferred for higher yield and better standardization. The following workflow outlines the key stages.
Materials:
- GMP-grade Collagenase (e.g., Collagenase NB6, Collagenase Type I, or Type II)
- Hyaluronidase (optional, for some protocols)
- Dulbecco's Modified Eagle Medium (DMEM)
- Fetal Bovine Serum (FBS) or Human Platelet Lysate (hPL)
- Antibiotic-Antimycotic solution
- Cell strainers (70-100 μm)
Procedure:
- Sample Pre-processing: Begin with steps identical to the explant protocol: rinse the cord, remove vessels, and mince the Wharton's jelly into 1-4 mm³ fragments [30].
- Enzymatic Digestion: Transfer the minced tissue to a tube containing the pre-warmed enzymatic solution. An optimized digestion condition uses 0.4 PZ U/mL of Collagenase NB6 for 3 hours at 37°C [30]. Alternative protocols use 0.2% Collagenase II for 16-20 hours [33]. Agitate the mixture periodically.
- Reaction Neutralization and Filtration: After digestion, neutralize the enzyme by adding a double volume of complete medium (e.g., DMEM with 10% FBS). Pass the resulting cell suspension through a 70-100 μm cell strainer to remove undigested tissue debris [33].
- Cell Pellet Collection: Centrifuge the filtrate at 1500 rpm for 10 minutes. Carefully discard the supernatant and resuspend the cell pellet in fresh complete culture medium [33].
- Primary Culture Initiation: Seed the cells at an appropriate density. For pilot-scale production, a seeding density of 0.5-2 grams of original tissue per 75 cm² flask is effective [30]. Incubate the culture at 37°C with 5% CO₂.
- Medium Change and Expansion: Perform the first medium change after 72-96 hours to remove non-adherent cells. Thereafter, change the medium every 2-3 days. Passage the cells upon reaching 70-80% confluence.
The Scientist's Toolkit: Essential Reagents and Materials
The following table lists key reagents required for the isolation of WJ-MSCs, with a focus on GMP-compliant options for translational research.
Table 3: Essential Research Reagent Solutions for WJ-MSC Isolation
| Reagent/Material | Function/Application | Examples & Notes |
|---|---|---|
| Collagenase Enzymes | Digests collagen in the extracellular matrix of Wharton's jelly to release cells. | Collagenase NB6 (GMP-grade): Recommended for clinical-grade production [30]. Collagenase Type I/II: Common for research; a modified protocol uses 0.2% Collagenase II [33]. |
| Culture Media | Provides nutrients and environment for cell growth and expansion. | DMEM/F12 or α-MEM are common basal media [31] [34]. |
| Serum Supplements | Provides essential growth factors and adhesion proteins. | Fetal Bovine Serum (FBS): 10-15% standard supplement [31]. Human Platelet Lysate (hPL): Xeno-free alternative; 2-5% concentration can be as effective as higher doses for expansion [30]. |
| Growth Factor Supplements | Enhances proliferation of MSCs. | Basic Fibroblast Growth Factor (bFGF): Adding 10 ng/mL bFGF significantly increases cell proliferation rates [31] [34]. |
| Antibiotic-Antimycotics | Prevents bacterial and fungal contamination in primary cultures. | Penicillin-Streptomycin (100-200 U/mL) is commonly used [31]. |
The selection between explant and enzymatic protocols depends heavily on the research or production objectives. The explant method is advantageous for its simplicity, lower cost, and reduced risk of introducing biological contaminants via enzymes, making it suitable for initial laboratory studies or settings with limited resources [29] [34]. However, its longer timeline and lower initial yield can be limiting.
Conversely, the enzymatic digestion method offers superior initial cell yield, faster initiation of culture, and, crucially, a more robust and standardized process, which is a fundamental requirement for Good Manufacturing Practice (GMP) compliance and scaling up to clinical-grade production [30] [32]. While it requires more optimization and carries a higher cost and theoretical risk of cell damage, modern GMP-compliant protocols have minimized these drawbacks [30].
In conclusion, both explant and enzymatic digestion are valid and effective for isolating WJ-MSCs. For large-scale, reproducible, and clinically-oriented manufacturing, an optimized enzymatic protocol is the definitive choice. Researchers should consider their specific needs for yield, time, reproducibility, and regulatory compliance when choosing their isolation pathway. Future work will continue to refine these protocols, particularly in scaling up production and further characterizing the subtle phenotypic differences that may exist between cells isolated via these distinct methods.
Within the broader scope of a thesis on mesenchymal stromal cell (MSC) isolation, this document provides detailed, source-specific enzymatic digestion protocols for two clinically relevant tissues: dental pulp and synovial membrane. The unique structural and compositional properties of each source tissue necessitate significant modifications to a generalized enzymatic digestion approach. Standardizing these protocols is critical for obtaining high yields of viable, functionally competent MSCs for downstream applications in regenerative medicine and drug development. This paper outlines these optimized protocols, complete with quantitative data, reagent toolkits, and standardized workflows to ensure experimental reproducibility.
Dental Pulp Stem Cell (DPSC) Isolation Protocol
The dental pulp is encapsulated within a mineralized tooth structure, requiring specialized mechanical access before enzymatic digestion can proceed. The protocol must minimize mechanical and chemical trauma to preserve the native stem cell population [35] [36].
Materials and Reagents
Table 1: Key Research Reagent Solutions for DPSC Isolation
| Reagent | Function / Purpose | Example / Concentration |
|---|---|---|
| Collagenase Type I | Enzymatic digestion of pulp extracellular matrix [36]. | 1 mg/mL from Clostridium histolyticum [36]. |
| Dulbecco's Modified Eagle's Medium (DMEM) | Base transport and culture medium [37] [35] [36]. | DMEM-Low Glucose [36]. |
| Fetal Bovine Serum (FBS) | Supplements culture medium; promotes cell adhesion and proliferation [35] [36]. | 10-20% (v/v) [35] [36]. |
| Antibiotic-Antimycotic | Prevents bacterial and fungal contamination [37] [35]. | e.g., Penicillin/Streptomycin & Amphotericin-B [37]. |
| Phosphate Buffered Saline (PBS) | Washing and diluting medium; maintains osmotic balance [35] [36]. | N/A |
| Cell Strainers | Filters digested tissue to obtain a single-cell suspension [37] [36]. | 40 μm, 100 μm, and 35 μm filters [37] [36]. |
Detailed Step-by-Step Method
- Tissue Acquisition and Transport: Obtain sound human third molars (e.g., extracted for orthodontic reasons) with informed consent and ethical approval. Teeth should be transported in a cold buffer (e.g., PBS) supplemented with high concentrations of antibiotics (e.g., 100 U/mL penicillin, 100 μg/mL streptomycin) [37] [35].
- Pulp Chamber Access: To avoid the mechanical trauma and heat generation of high-speed dental drills, use a controlled method to access the pulp chamber. One recommended approach is using a water-cooled diamond blade saw to make precise grooves around the tooth, which can then be split with a sterilized chisel and mallet [35]. This method avoids direct contact with the pulp tissue itself.
- Pulp Tissue Extraction: Gently remove the intact pulp tissue using sterile forceps or a curette [36].
- Washing and Mincing: Transfer the pulp to a Petri dish and wash twice with DMEM containing antibiotics [37]. Using sterile surgical scissors or a scalpel, mince the tissue into small fragments of approximately 1–2 mm² [37].
- Enzymatic Digestion: Transfer the minced tissue to a conical tube containing a collagenase solution (e.g., 1 mg/mL in PBS) [36]. Incubate in a water bath at 37°C for 45-60 minutes with continuous agitation [36].
- Digestion Neutralization and Filtration: Neutralize the enzyme action by adding a complete culture medium containing FBS. Pass the resulting cell suspension sequentially through 100 μm and 35 μm cell strainers to remove undigested tissue fragments and obtain a single-cell suspension [36].
- Centrifugation and Seeding: Centrifuge the filtered suspension at 1800 rpm for 10 minutes at 4°C [36]. Resuspend the cell pellet in a complete culture medium (e.g., DMEM supplemented with 10-20% FBS and antibiotics) and seed into culture flasks [35] [36].
Figure 1: Experimental workflow for the isolation of Dental Pulp Stem Cells (DPSCs) from a third molar.
Synovial Membrane Cell Isolation Protocol
The synovial membrane is a soft, vascularized tissue lining joint cavities. Its dissociation requires a robust enzymatic cocktail to break down the matrix and release embedded fibroblasts and macrophages efficiently [38] [39].
Materials and Reagents
Table 2: Key Research Reagent Solutions for Synovial Cell Isolation
| Reagent | Function / Purpose | Example / Concentration |
|---|---|---|
| Liberase TL | Enzyme blend (Collagenase I/II + neutral protease) for gentle and effective tissue dissociation [38] [39]. | 100 μg/mL [38]. |
| DNase I | Degrades neutrophil extracellular traps (NETs) and free DNA released during digestion, reducing clumping and increasing cell yield [38]. | 100 μg/mL [38]. |
| Antibodies for FACS | Identification and isolation of specific cell populations (e.g., macrophages, fibroblasts) [38]. | Anti-CD45, Anti-CD14, Anti-PDPN [38]. |
| Fluorescence-Activated Cell Sorter (FACS) | High-purity isolation of specific synovial cell subtypes based on surface marker expression [38]. | N/A |
Detailed Step-by-Step Method
- Tissue Acquisition and Confirmation: Obtain synovial tissue biopsies from patients undergoing procedures like hip arthroscopy or joint replacement. The anterior synovial fold during hip arthroscopy is a confirmed viable location for harvest [38]. A small piece of tissue should be fixed for cryosection to confirm typical synovial histology (lining and sublining layers) [38].
- Washing and Mincing: Wash the tissue in PBS to remove blood and debris. Mince the tissue into the finest possible pieces using sterile surgical blades [38].
- Optimized Enzymatic Digestion: Transfer the minced tissue to a tube containing the optimized enzyme cocktail: 100 μg/mL Liberase TL and 100 μg/mL DNase I in PBS [38] [39]. Incubate at 37°C for 60-90 minutes with continuous agitation.
- Digestion Neutralization and Filtration: Neutralize the enzymatic activity by adding a complete culture medium with serum. Pass the cell suspension through a 70 μm or 100 μm cell strainer to remove debris and obtain a single-cell suspension.
- Cell Washing and Counting: Centrifuge the filtered suspension and resuspend the pellet in a suitable buffer (e.g., FACS buffer). Perform a cell count and viability assessment using Trypan Blue exclusion.
- Cell Sorting and Characterization (Optional): For isolation of specific cell types, incubate the cell suspension with antibodies against established markers:
Figure 2: Experimental workflow for the isolation of cells from the synovial membrane, including an optional FACS sorting step.
Critical Parameter Comparison and Troubleshooting
A direct comparison of the optimized parameters for each tissue source highlights the necessity for protocol customization.
Table 3: Quantitative Protocol Parameter Comparison
| Parameter | Dental Pulp Protocol | Synovial Membrane Protocol |
|---|---|---|
| Primary Enzyme | Collagenase Type I (1 mg/mL) [36] | Liberase TL (100 μg/mL) [38] |
| Supplemental Enzyme | Not typically used | DNase I (100 μg/mL) [38] |
| Digestion Time | 45-60 minutes [36] | 60-90 minutes [38] [39] |
| Digestion Temperature | 37°C [36] | 37°C [38] |
| Key Filtration Steps | 100 μm → 35 μm [36] | 70-100 μm [38] |
| Critical Cell Markers | CD90, CD73, CD105, CD44 [40] | Fibroblasts: PDPN+/CD45- [38]Macrophages: CD45+/CD14+ [38] |
Common challenges in DPSC isolation include bacterial contamination, lack of cell adherence, and the presence of undissolved particles in the culture medium, which can be mitigated by strict aseptic technique, careful reagent selection, and proper filtration [36]. For synovial tissue, the key is using the Liberase TL + DNase I cocktail to maximize the yield of viable cells from small biopsy specimens for high-quality single-cell RNA sequencing [38] [39].
The enzymatic digestion of tissues is a critical initial step in isolating Mesenchymal Stromal Cells (MSCs). However, the subsequent processing steps—centrifugation, washing, and plating—are equally vital for determining the yield, purity, and functional characteristics of the final cell product. Within the broader context of developing robust enzymatic digestion protocols for MSC isolation, standardizing these post-digestion procedures is essential for reducing technical variability and enhancing the comparability of research and clinical outcomes [27]. This protocol details evidence-based strategies for these crucial steps, drawing on optimized methods from recent studies across various tissue sources.
Core Post-Digestion Workflow
The fundamental process following enzymatic digestion involves separating the released stromal vascular fraction (SVF) from debris, residual enzymes, and contaminants, followed by the establishment of primary cultures. The diagram below illustrates the standard workflow and key decision points.
Detailed Protocols and Methodologies
Centrifugation Strategies
The primary goal of centrifugation is to pellet the mononuclear cell fraction while eliminating erythrocytes, cellular debris, and neutralized enzymes.
Standard Density Gradient Centrifugation: This method is well-established for isolating MNCs from bone marrow and other tissues [41]. The process involves carefully layering the diluted cell suspension onto a density gradient medium (e.g., Ficoll-Paque PLUS) in a tube. Centrifugation is typically performed at 300-400 ×g for 30 minutes at room temperature (21°C) without brakes to prevent disturbance of the separated layers [41]. After centrifugation, the opaque MNC ring at the plasma-Ficoll interface is carefully aspirated and transferred to a new tube for subsequent washing.
Direct Centrifugation without Gradient: For tissues like adipose or umbilical cord that have been enzymatically digested, direct centrifugation is often sufficient. The protocol from thymic tissue isolation specifies two sequential spins: a first centrifugation at 400 ×g for 10 minutes at 4°C to pellet the SVF, followed by a second spin at 400 ×g for 5 minutes at 20°C after resuspension to remove residual collagenase [42]. This method effectively separates buoyant adipocytes and tissue debris from the pelleted SVF.
Washing Procedures
Washing is critical for removing enzymes that could damage cells during culture and for eliminating contaminants that impair cell adherence and growth.
Protocol:
- Resuspend Pellet: After centrifugation, carefully aspirate the supernatant and resuspend the cell pellet in a generous volume (e.g., 10-50 mL depending on initial volume) of wash medium.
- Wash Medium Composition: Use a basic medium such as Dulbecco's Phosphate Buffered Saline (DPBS) or α-Minimum Essential Medium (α-MEM). The medium is often supplemented with 2-20% Fetal Bovine Serum (FBS) or human platelet lysate to neutralize trypsin and protect cells, and with antibiotics (e.g., Penicillin/Streptomycin) to prevent contamination [43] [41] [42].
- Repeat Centrifugation: Centrifuge the resuspended cells at 400-1200 ×g for 5-10 minutes [43] [42].
- Final Resuspension: Aspirate the supernatant and resuspend the final pellet in a complete culture medium suitable for plating. A final filtration step through 100 µm and/or 40 µm cell strainers is recommended to remove cell clumps and tissue aggregates, ensuring a single-cell suspension for accurate counting and uniform plating [42].
Plating Strategies for Primary Culture
The initial plating strategy is a key determinant for successful MSC expansion, influencing both the initial adherence of the target cell population and the overall culture purity.
Standard Plating Protocol:
- Cell Counting: Perform a cell count using an automated cell counter (e.g., Sysmex XN-20) or hemocytometer to determine the total number of viable nucleated cells.
- Seeding Density: Seed the cells at an appropriate density based on the source and isolation method.
- For bone marrow MNCs, a high density of 160,000 cells/cm² is used to maximize the chance of MSC adherence and colony formation [41].
- For adherent SVF cells from tissues like adipose or thymus, plating the entire SVF pellet from the digestion or seeding at densities around 2.5 × 10⁵ cells/cm² is effective [42].
- Culture Conditions: Plate cells on untreated plastic culture flasks or dishes in a complete growth medium. Incubate at 37°C in a humidified atmosphere of 5% CO₂ [43] [41] [42].
- Medium Refreshment: The first complete medium change should be conducted 24 hours post-plating to remove non-adherent cells, primarily hematopoietic lineages. This significantly enhances the purity of the resulting MSC culture. Subsequent medium changes are typically performed every 2-3 days.
Quantitative Data and Comparisons
Impact of Isolation Method on Cell Yield and Characteristics
Table 1: Comparison of MSC Yields and Characteristics from Different Tissues and Isolation Methods
| Tissue Source | Isolation Method | Key Yield/Characteristic Findings | Study Details |
|---|---|---|---|
| Bone Marrow | Automated (Sepax) vs. Manual Ficoll | Automated method yielded slightly higher MNCs; No significant difference in subsequent CFU formation or MSC characteristics. [41] | 17 donor samples; GMP conditions. |
| Adipose Tissue | Enzymatic (SVF) vs. Mechanical (MF) | Both methods yielded MSCs with typical characteristics; Secretome composition varied significantly based on extraction method. [43] | 4 donors; Secretome analysis. |
| Human Umbilical Cord | Explant Culture | High robustness: 98.9% purity, >97% viability. Vessel removal before explant culture improved MSC purity. [27] | 90 UC donors; Standardized protocol. |
| Adult Thymus | Enzymatic vs. Outgrowth | Both methods yielded fibroblast-like, plastic-adherent cells with high proliferation and trilineage potential. [42] | Xeno-free culture with human platelet lysate. |
Reagent Solutions for Post-Digestion Processing
Table 2: Essential Reagents and Materials for Post-Digestion Processing
| Reagent/Material | Function/Application | Example Specifications |
|---|---|---|
| Ficoll-Paque PLUS | Density gradient medium for the separation of mononuclear cells from other cellular components. [41] | Centrifuge at 300-400 ×g for 30 min. |
| Centrifuge Tubes | For containing samples during centrifugation and density gradient separation. | 50 mL conical tubes (e.g., Corning). [41] |
| Wash/Base Medium | Diluent and washing solution to neutralize enzymes and remove debris. | α-MEM or DMEM. [41] [42] |
| Serum Supplement | Provides essential nutrients and growth factors; inactivates residual trypsin. | Fetal Bovine Serum (FBS, 10-20%) or human Platelet Lysate (pHL, 10%). [41] [42] |
| Antibiotic-Antimycotic | Prevents bacterial and fungal contamination in primary cultures. | Penicillin (100 U/mL)/Streptomycin (0.1 mg/mL). [43] [42] |
| Cell Strainers | Removal of cell clumps and tissue aggregates to generate a single-cell suspension. | 100 µm and 40 µm mesh sizes. [42] |
| Culture Flasks/Plates | Substrate for cell adhesion and expansion. | Untreated plastic (e.g., 175 cm² flasks). [41] |
Critical Factors for Success and Troubleshooting
- Time Sensitivity: Processing delays can impact cell viability. Initiate processing of human umbilical cord samples within 6 hours of birth, and complete UC processing within 48 hours of collection for optimal results [27].
- Serum Selection: The choice of serum can influence expansion reproducibility. Compared to FBS, human platelet lysate has been shown to increase the reproducibility of expansion rates and MSC characteristics [27].
- Handling of Explants: When using the explant method (e.g., for umbilical cord or thymus), removing blood vessels before initiating explant cultures is a critical step to improve the purity of the resulting MSC culture [27].
- Quality Control: Always assess the quality of the initial cell suspension post-washing. This includes cell counts, viability assays (e.g., Trypan Blue exclusion), and visual inspection for excessive red blood cell contamination, which may require additional purification steps.
The manufacture of Advanced Therapy Medicinal Products (ATMPs), which includes therapies based on mesenchymal stromal cells (MSCs), is governed by a stringent regulatory framework to ensure product quality, safety, and efficacy. Under European Union law, the manufacture or import of any medicinal product requires a manufacturing authorization, and production must comply with Good Manufacturing Practice (GMP) principles [44]. For ATMPs, the European Commission has established specific guidelines outlined in EudraLex Volume 4, Part IV, which details GMP requirements tailored to the unique complexities of advanced therapies [45] [44]. These guidelines are mandatory for ATMPs that have received a marketing authorization and are also applied in the clinical trial setting [44].
The core principle of GMP is a comprehensive Pharmaceutical Quality System, which encompasses the total sum of organized arrangements to ensure medicinal products possess the quality required for their intended use [44]. For ATMPs, this is particularly critical due to their inherent complexities, such as the use of substances of human origin (like tissues and cells for MSC isolation), limited potential for sterile filtration, and, in the case of personalized therapies, the challenge of manufacturing many small batches for individual patients [45]. The regulatory landscape is dynamic, with the European Medicines Agency (EMA) proposing revisions to Part IV in 2025 to align with updated standards and technological advancements [46] [47].
Current EU GMP Guidelines for ATMPs
Key Regulatory Documents and Oversight
The primary GMP framework for ATMPs in the European Union is defined by the following key documents and oversight bodies:
- EudraLex Volume 4, Part IV: This is the central guideline on GMP specific to ATMPs. Published in 2017, it provides a stand-alone set of standards that acknowledges the distinct challenges of ATMP manufacturing, emphasizing a risk-based approach (RBA) [45] [44]. It diverges from the approach for other biologics and states that other annexes of EudraLex Volume 4 are not applicable to ATMPs [45].
- PIC/S Annex 2A: The Pharmaceutical Inspection Co-operation Scheme (PIC/S) has taken a different approach by integrating ATMP requirements into its GMP annexes. Its Annex 2A applies to ATMPs and frequently references the principles of Annex 1 (Manufacture of Sterile Medicinal Products), creating a potential point of divergence from the EU's Part IV [45].
- Annex 1 (Manufacture of Sterile Medicinal Products): Although Part IV is designed as a stand-alone document, the updated Annex 1, which became effective in August 2023, has significant implications for aseptic processing. Many ATMP manufacturers choose to apply its principles, especially for processes that cannot undergo sterile filtration [46] [45].
- Competent Authorities: National competent authorities in EU Member States are responsible for assessing applications for manufacturing authorizations and conducting regular on-site inspections to verify GMP compliance. The European Medicines Agency (EMA) plays a coordinating role, and the GMP/GDP Inspectors Working Group provides harmonized guidance across the European Economic Area [44].
Table 1: Key GMP Regulatory Documents for ATMPs
| Document | Issuing Body | Scope & Relevance to ATMPs | Status |
|---|---|---|---|
| EudraLex Vol. 4, Part IV | European Commission | Stand-alone GMP guidelines for ATMPs; emphasizes risk-based approach. | Currently in force; revision proposed in 2025 [46]. |
| PIC/S Annex 2A | Pharmaceutical Inspection Co-operation Scheme (PIC/S) | GMP requirements for ATMPs within the annex structure; references Annex 1. | In force, creating some divergence from EU Part IV [45]. |
| Annex 1 | European Commission / PIC/S | Principles for manufacturing sterile products; highly relevant for aseptic ATMP processes. | Updated version effective August 2023 [46] [45]. |
Proposed Revisions to the Regulatory Framework
The regulatory framework for ATMP GMP is continuously evolving. In May 2025, the EMA released a concept paper proposing a revision of Part IV [46] [47]. The driving forces behind this proposed update include:
- Alignment with Revised Annex 1: The updated Annex 1 introduced modifications for the manufacture of sterile medicinal products. The revision seeks to harmonize ATMP-specific GMP requirements with these changes, particularly emphasizing a Contamination Control Strategy (CCS) [46].
- Integration of ICH Guidelines: The revision plans to incorporate principles from ICH Q9 (Quality Risk Management) and ICH Q10 (Pharmaceutical Quality System), promoting a more systematic approach to risk management and quality systems [46].
- Adaptation to Technological Advancements: The guidelines will provide clarifications on qualifying and controlling new technologies, such as automated systems, closed single-use systems, and rapid microbiological testing methods [46].
- Updates on Cleanroom and Barrier Systems: Further clarifications are expected regarding cleanroom classifications and the use of isolators and Restricted Access Barrier Systems (RABS), while maintaining provisions for biosafety cabinets used in manual manipulations [46].
The public consultation for this revision was open until July 2025, indicating an ongoing effort to refine the regulatory environment for ATMPs [46].
GMP-Compliant MSC Isolation: From Protocol to Production
The isolation of Mesenchymal Stromal Cells (MSCs) is a critical first step in the manufacturing of many ATMPs. GMP requires that this process is robust, validated, and controlled to ensure a consistent and high-quality cell source.
MSC Isolation Techniques and Comparative Analysis
The two predominant methods for isolating MSCs from tissues like adipose tissue, bone marrow, or umbilical cord are enzymatic digestion and the explant culture method [48]. The choice of method impacts cell yield, viability, functionality, and compliance with GMP principles.
Table 2: Comparison of MSC Isolation Techniques
| Parameter | Enzymatic Digestion | Explant Culture Method |
|---|---|---|
| Basic Principle | Uses proteolytic enzymes (e.g., Collagenase, Liberase) to digest the extracellular matrix and release individual cells [12] [48]. | Tissue pieces are plated directly, allowing cells to migrate out from the explant and adhere to the culture surface [48]. |
| Process Speed | Rapid; cells typically reach confluence within ~7 days [48]. | Slower; can take up to ~10-15 days for cells to reach confluence [8] [48]. |
| Cell Yield | Generally high cell yield, but dependent on enzyme and protocol [8]. | Lower initial cell yield compared to enzymatic methods [8]. |
| Cell Viability & Function | Risk of cell damage due to over-digestion; may require growth factors to support primary culture [48]. | Better preservation of cell integrity; shorter population doubling times and maintained genetic stability [48]. |
| GMP Considerations | Requires validated enzyme specifications and digestion parameters; better suited for scalable, closed-system automation. | Avoids introduction of enzymatic reagents; simpler process but may be less scalable and more prone to operator variation. |
Optimized Enzymatic Digestion Protocol for Adipose-Derived MSCs (AD-MSCs)
The following protocol provides a detailed, GMP-oriented methodology for the enzymatic isolation of AD-MSCs, based on current research. This protocol can serve as a template for developing and validating a Standard Operating Procedure (SOP).
Objective: To isolate viable AD-MSCs from solid adipose tissue or lipoaspirate for use in GMP-compliant ATMP manufacturing.
Materials and Reagents:
- Sterile Phosphate-Buffered Saline (PBS)
- Type I Collagenase or Liberase (GMP-grade)
- Complete Growth Medium (e.g., α-MEM supplemented with fetal bovine serum or human platelet lysate, and antibiotics)
- Cell Strainers (70µm or 100µm)
- Centrifuge Tubes
Procedure:
- Tissue Collection and Transport: Aseptically collect adipose tissue (e.g., subcutaneous fat) in a sterile container with a transport medium designed to maintain tissue viability. Document the tissue source and donor information as per GMP traceability requirements [44].
- Washing: Transfer the adipose tissue to a sterile Petri dish. Wash the tissue thoroughly with PBS to remove blood contaminants and residual local anesthetics [12].
- Enzymatic Digestion: a. Mince the washed adipose tissue into small pieces (~2-4 mm³) using sterile scalpels or scissors. b. Transfer the minced tissue to a digestion vessel and add a pre-warmed GMP-grade enzyme solution. Research indicates that using 0.1% Liberase for 3 hours under controlled temperature (37°C) and agitation provides a high cell yield with low population doubling time [8]. c. Incubate the mixture with continuous agitation for the validated duration.
- Digestion Neutralization and Centrifugation: a. After digestion, neutralize the enzyme activity by adding an equal volume of complete growth medium [12]. b. Centrifuge the mixture at a low speed (e.g., 300-600 x g for 10 minutes) to separate the cellular pellet, known as the Stromal Vascular Fraction (SVF), from the adipocytes and residual tissue debris [12].
- Cell Pellet Resuspension and Filtration: a. Carefully aspirate the supernatant, including the floating adipocyte layer and medium. b. Resuspend the cell pellet in fresh complete growth medium. c. Filter the cell suspension through a cell strainer (e.g., 100µm) to remove any residual tissue fragments and obtain a single-cell suspension [12].
- Seeding and Primary Culture: a. Count the cells and assess viability (e.g., using Trypan Blue exclusion). b. Seed the cells at a validated density into culture vessels pre-treated with a GMP-compliant cell attachment surface enhancer (e.g., Corning CellBIND) to improve attachment and yield [48]. c. Incubate the culture at 37°C in a humidified atmosphere with 5% CO₂. d. Refresh the culture medium every 2-3 days to remove non-adherent cells.
- Cell Passaging and Banking: a. Passage the cells when they reach 70-80% confluence, typically using a GMP-grade detachment reagent. b. Establish a Master Cell Bank (MCB) and a Working Cell Bank (WCB) following GMP banking principles to ensure a consistent and traceable cell source for production [48].
Quantitative Evaluation of Enzymatic Conditions
Optimizing enzymatic digestion is critical for maximizing yield in a GMP context. A 2024 study systematically evaluated 32 different conditions for isolating bovine AD-MSCs, providing a quantitative basis for protocol selection [8].
Table 3: Evaluation of Enzymatic Conditions for Bovine AD-MSC Isolation [8]
| Enzyme | Concentration | Incubation Time | Average Cell Yield (x10⁶ cells/g tissue) | Key Findings |
|---|---|---|---|---|
| Collagenase Type I (Coll IA) | 0.1% | 3 h | Reference | Most frequently reported enzyme for AT-MSC isolation. |
| Collagenase Type I (Coll IA) | 0.1% | 6 h | > 35 | Successful isolation, selected for further characterization. |
| Collagenase Type I (Coll IA) | 0.04% | 6 h | > 35 | Successful isolation, selected for further characterization. |
| Liberase (LibTM) | 0.1% | 3 h | 30.48 - 67.1 | Significantly higher yield vs. Coll IA 0.1% 3h; fastest proliferation. |
| Liberase (LibTM) | 0.1% | 6 h | > 35 | Successful isolation, selected for further characterization. |
| Liberase (LibTM) | 0.1% | Overnight | > 35 | Successful isolation, selected for further characterization. |
| Collagenase Type IV (Coll IV) | 0.04% | 6 h, ON, 24 h | 0 | No plastic-adherent cells observed. |
Integration of GMP Principles in MSC Isolation Workflows
Implementing GMP for MSC-based ATMPs requires a holistic, risk-based approach that integrates quality controls throughout the entire isolation and manufacturing process. The following workflow diagram synthesizes the technical protocol with the overarching GMP framework.
GMP-Compliant MSC Isolation Workflow
The Scientist's Toolkit: Essential Reagents and Materials
The following table details key reagents and materials required for a GMP-compliant MSC isolation process, along with their critical functions and quality considerations.
Table 4: Research Reagent Solutions for GMP-Compliant MSC Isolation
| Reagent/Material | Function in Protocol | GMP & Quality Considerations |
|---|---|---|
| Liberase or Collagenase (GMP-grade) | Proteolytic digestion of the extracellular matrix to release the Stromal Vascular Fraction (SVF) [8]. | Use of GMP-grade, well-characterized enzymes is critical. Certificate of Analysis (CoA) required for identity, purity, and potency [44]. |
| Cell Culture Media & Supplements | Provides nutrients and factors for cell growth, expansion, and maintenance [48]. | Preference for defined, xeno-free formulations. All components (e.g., FBS alternatives like human platelet lysate) must be qualified and tested for adventitious agents [44]. |
| CellBIND or Enhanced Surfaces | Culture surface treatment that enhances cell attachment and improves initial yield [48]. | Materials must be qualified for use. Single-use, pre-sterilized vessels can reduce cleaning validation and cross-contamination risks. |
| Sterile Single-Use Assemblies | For fluid transfer, filtration, and containment during processing [45]. | Enables a closed processing strategy, which is a key element of the contamination control strategy, potentially allowing for lower cleanroom classification [45]. |
| Validated Detection Kits | For in-process controls like cell counting, viability, and sterility testing. | Methods must be validated for their intended use. Rapid microbiological methods are encouraged to support faster batch release [46]. |
The successful development and manufacturing of MSC-based ATMPs hinge on the seamless integration of robust, optimized scientific protocols with a rigorous GMP framework. The enzymatic isolation of MSCs, as detailed in these application notes, must be designed with scalability, consistency, and control in mind. The ongoing evolution of guidelines, such as the proposed revision of EudraLex Volume 4, Part IV, underscores the importance of a proactive and flexible risk-based approach that can adapt to new technologies and scientific understanding. By adhering to these principles—employing validated reagents, implementing comprehensive in-process controls, establishing well-characterized cell banks, and maintaining a robust Pharmaceutical Quality System—researchers and drug development professionals can navigate the complex regulatory landscape and advance promising MSC therapies from the laboratory to the clinic.
Maximizing Yield and Viability: A Troubleshooting and Optimization Guide
The isolation of Mesenchymal Stromal Cells (MSCs) through enzymatic digestion of tissues is a critical first step in producing cell-based therapies. The efficiency of this process directly impacts cell yield, viability, proliferative capacity, and ultimately, the therapeutic potential of the final product. The optimization of key parameters—enzyme concentration, incubation time, and temperature—is therefore paramount for developing robust, reproducible, and scalable manufacturing protocols compliant with Good Manufacturing Practice (GMP) standards [49]. This Application Note synthesizes current research to provide detailed, evidence-based guidance on optimizing these critical parameters across different tissue sources, including adipose tissue, umbilical cord, and bone marrow, for clinical-grade MSC production.
Research consistently demonstrates that the interplay between enzyme type, concentration, and incubation time significantly influences MSC isolation outcomes. The following table summarizes optimized parameters from recent studies for different tissue sources.
Table 1: Optimized Enzymatic Digestion Parameters for MSC Isolation from Various Tissues
| Tissue Source | Optimal Enzyme | Optimal Concentration | Optimal Incubation Time | Key Outcomes | Source/Study Context |
|---|---|---|---|---|---|
| Bovine Adipose Tissue | Liberase | 0.1% | 3 hours | Highest cell yield with low population doubling time; confirmed MSC identity and myogenic potential. | [8] |
| Human Umbilical Cord (Wharton's Jelly) | Collagenase NB6 (GMP-grade) | 0.4 PZ U/mL | 3 hours | Higher yield of P0 WJ-MSCs; established as part of a scalable GMP-compliant manufacturing process. | [50] |
| Human Infrapatellar Fat Pad | Collagenase I | 0.1% | 2 hours | Standardized protocol for isolation; part of a feasibility study for GMP-compliant clinical use. | [51] |
| Equine Adipose Tissue | Collagenase I | 0.8 mg/mL | 4 hours | Significantly higher MSC yield compared to the explant method, with no major differences in proliferation or differentiation. | [52] |
The Impact of Parameter Interaction
Optimization is not achieved by considering parameters in isolation. A study on bovine adipose tissue systematically evaluated 32 conditions, combining four enzymes (Collagenase I, Collagenase I + Trypsin, Liberase, and Collagenase IV) with varying concentrations and incubation times [8]. The findings revealed that while varying concentration or time alone did not always yield statistically significant differences, higher cell yields were consistently observed when a 0.1% enzyme concentration was combined with shorter incubation times (3-6 hours). This underscores the importance of a balanced approach; excessive incubation times, even with lower enzyme concentrations, can compromise cell viability and functionality.
Detailed Experimental Protocols
Protocol: Optimization of Enzymatic Digestion for Bovine Adipose Tissue-Derived MSCs
This protocol is adapted from a study that identified Liberase as the most effective enzyme for isolating MSCs from bovine subcutaneous adipose tissue [8].
3.1.1 Reagents and Materials
- Liberase (Research Grade)
- Collagenase Type I (for comparative conditions)
- Dulbecco's Phosphate-Buffered Saline (DPBS), without Ca2+/Mg2+
- Complete culture medium (e.g., α-MEM supplemented with fetal bovine serum and antibiotics)
- Sterile surgical scalpels and forceps
- 70% ethanol for disinfection
- Water bath, calibrated to 37°C
- Centrifuge
- Cell strainer (70-100 µm)
3.1.2 Procedure
- Tissue Harvesting and Preparation: Aseptically collect subcutaneous adipose tissue. Rinse tissue thoroughly with DPBS containing 1% Penicillin-Streptomycin-Neomycin to remove residual blood. Mince the tissue into fine fragments (< 0.1-0.2 cm³) using sterile scalpels.
- Enzyme Solution Preparation: Prepare a working solution of 0.1% (w/v) Liberase in DPBS. Filter-sterilize the solution using a 0.22 µm filter.
- Digestion Process: Incubate the minced adipose tissue in the Liberase solution using a 1:1 (w/v) ratio. Place the mixture in a continuously shaking water bath at 37°C for 3 hours.
- Digestion Termination and Cell Harvesting: After incubation, neutralize the enzyme action by adding an equal volume of complete culture medium. Centrifuge the cell suspension at 300-400 × g for 10 minutes to pellet the stromal vascular fraction (SVF).
- Cell Washing and Seeding: Resuspend the cell pellet in DPBS, pass it through a 100 µm cell strainer to remove undigested tissue, and centrifuge again. Resuspend the final cell pellet in complete culture medium and seed at a density of approximately 20,000 cells/cm².
3.1.3 Validation and QC
- Cell Yield and Viability: Determine using a trypan blue exclusion assay. The optimized protocol should yield >35 × 10⁶ cells/g tissue at P1 with >95% viability [8].
- Phenotypic Characterization: Confirm MSC identity via flow cytometry for CD73, CD90, and CD105 positivity and CD45 negativity.
- Functional Potency: Validate tri-lineage differentiation potential (osteogenic, adipogenic, chondrogenic) and, if applicable, myogenic differentiation capacity.
Protocol: GMP-Compliant Isolation of Human Wharton's Jelly MSCs
This protocol outlines a standardized, scalable method for isolating clinical-grade MSCs from the human umbilical cord [50] [27].
3.2.1 Reagents and GMP-Compliant Materials
- GMP-grade Collagenase NB6
- DPBS (without Ca2+/Mg2+)
- Serum-free, xeno-free MSC culture medium (e.g., NutriStem)
- Human Platelet Lysate (hPL)
- 0.5% Povidone-iodine solution for decontamination
3.2.2 Procedure
- Umbilical Cord Collection and Pre-processing: Obtain umbilical cord (>20 cm length) post-cesarean section with informed consent. Transport at 2-10°C within 24 hours. Rinse with DPBS and decontaminate with 0.5% povidone-iodine for 3 minutes, followed by three DPBS rinses.
- Wharton's Jelly Dissection: Dissect the cord to expose Wharton's jelly, carefully remove the two arteries and one vein. Mince the remaining Wharton's jelly into 1-4 mm³ fragments.
- Enzymatic Digestion: Digest the tissue fragments using 0.4 PZ U/mL Collagenase NB6 in a shaking incubator at 37°C for 3 hours.
- Cell Seeding and Expansion: After digestion, neutralize with culture medium, filter, and centrifuge to pellet cells. Seed the cells in a T-75 flask using serum/xeno-free medium supplemented with 2-5% hPL.
3.2.3 Process Validation
- Robustness: The method should be robust across donors, with UCs processed up to 48 hours post-collection without impacting MSC characteristics [27].
- Characterization: Isolated MSCs should show >97% viability, ~98.9% purity (by surface marker expression), and functional immunomodulatory capacity in T-cell proliferation assays.
Workflow Visualization
The following diagram illustrates the logical sequence and decision points involved in optimizing the critical parameters for MSC isolation via enzymatic digestion.
Diagram 1: Parameter optimization workflow for MSC isolation.
The Scientist's Toolkit: Essential Research Reagent Solutions
Successful and reproducible isolation of MSCs hinges on the selection of appropriate, high-quality reagents. The following table details key materials and their functions.
Table 2: Essential Reagents for Enzymatic Digestion in MSC Isolation
| Reagent / Material | Function / Role in Protocol | Examples & Key Considerations |
|---|---|---|
| Proteolytic Enzymes | Breaks down the extracellular matrix to release individual cells from the tissue stroma. | Liberase: A purified enzyme blend effective for adipose tissue [8]. Collagenase NB6 (GMP): A GMP-compliant enzyme critical for clinical-grade UC-MSC production [50]. Collagenase I: Widely used for various tissues, including fat pad and equine tissues [51] [52]. |
| Basal Buffers | Provides an isotonic environment for tissue washing, enzyme preparation, and cell washing. | Dulbecco's PBS (without Ca²⁺/Mg²⁺): Prevents enzyme inhibition and cell clumping. |
| Culture Medium | Provides nutrients for cell growth post-isolation and is used to neutralize enzymatic activity. | Serum/Xeno-Free Media (e.g., NutriStem): Redances batch variability and safety risks for clinical applications [50] [51]. Media with Human Platelet Lysate (hPL): A human-derived alternative to FBS for GMP processes [50] [27]. |
| Decontamination Agents | Minimizes microbial contamination from the starting tissue. | 0.5% Povidone-iodine: Standard for decontaminating umbilical cords before processing [50]. Antibiotic/Antimycotic Solutions (e.g., Gentamicin): Added to wash and initial culture media to prevent contamination [52]. |
Mesenchymal stromal cell (MSC) research continues to advance regenerative medicine, with promising applications across diverse fields including inflammatory bowel disease treatment, osteoarthritis therapy, and cultured meat production [53] [54] [8]. Despite this potential, the transition from laboratory research to clinically viable therapies faces significant manufacturing challenges. Three persistent obstacles—low cell yield, poor cell viability, and microbial contamination—consistently compromise product quality, safety, and efficacy. These issues are particularly pronounced during the critical initial phase of MSC isolation via enzymatic digestion from various tissue sources such as adipose tissue, bone marrow, and umbilical cord. This application note provides a structured framework to identify, troubleshoot, and resolve these common yet critical problems through optimized protocols, quantitative comparisons, and rigorous quality control measures, ultimately supporting more robust and reproducible MSC manufacturing for therapeutic applications.
Troubleshooting Guide: Identification and Resolution of Common Issues
The following section outlines a systematic approach to diagnosing and addressing the most frequent challenges in MSC isolation protocols. The table below summarizes key issues, their potential causes, and evidence-based solutions.
Table 1: Troubleshooting Guide for MSC Isolation Protocols
| Problem | Potential Causes | Recommended Solutions |
|---|---|---|
| Low Cell Yield | Suboptimal enzyme selection, insufficient digestion time, low enzyme concentration, or inefficient tissue dissociation [8]. | Optimize enzyme cocktail (e.g., use 0.1% Liberase for 3 hours for adipose tissue); increase enzyme concentration or digestion time within validated limits; ensure proper tissue mincing [8]. |
| Poor Cell Viability | Excessive digestion time, overly high enzyme concentration, harsh mechanical manipulation, or improper neutralization of enzymes [8] [55]. | Shorten digestion time; titrate enzyme to lowest effective concentration; use gentler pipetting; ensure complete enzyme neutralization with serum-containing medium or inhibitors [8]. |
| Microbial Contamination | Non-sterile starting tissue, inadequate disinfection protocols, or failure in aseptic processing techniques [54] [49]. | Implement rigorous donor site disinfection and tissue decontamination (e.g., antibiotic washes); validate cleanroom practices with monitoring systems like AMIL link technology [54]. |
Quantitative Comparison of Isolation Methods and Outcomes
Selecting the appropriate isolation method is fundamental to achieving high-quality MSC products. The table below provides a comparative analysis of enzymatic and mechanical approaches based on key performance metrics, synthesizing data from recent studies.
Table 2: Performance Comparison of MSC Isolation Methods
| Isolation Method | Average Cell Yield | Cell Viability | Key Markers/Properties | Primary Advantages & Disadvantages |
|---|---|---|---|---|
| Enzymatic (Adipose, 0.1% Liberase, 3h) [8] | 30.48 - 67.1 x 106 cells/g tissue | >95% | CD73+, CD90+, CD105+; Tri-lineage differentiation potential [8]. | Advantages: High cell yield, effective tissue dissociation, reproducibility.Disadvantages: Higher cost, regulatory considerations ("more than minimal manipulation") [55]. |
| Mechanical (Adipose SVF) [55] | ~11.96 x 104 cells/ml lipoaspirate | Up to 98% | CD34+ (8.70%), CD73+ (11.63%), CD105+ (4.08%); Retained immunomodulatory function [55]. | Advantages: Faster, avoids enzymatic reagents, favorable regulatory status.Disadvantages: Lower total cell yield, potential for lower purity [55]. |
Experimental Workflow for Optimized MSC Isolation
The following diagram and detailed protocol describe an optimized workflow for the enzymatic isolation of MSCs from adipose tissue, incorporating steps to maximize yield and viability while preventing contamination.
Diagram Title: Optimized MSC Isolation and Quality Control Workflow
Detailed Protocol: Enzymatic Isolation of Adipose-Derived MSCs (AD-MSCs)
Principle: This protocol utilizes controlled enzymatic digestion to release the stromal vascular fraction (SVF) from adipose tissue, followed by plating to isolate the plastic-adherent MSC population [12].
Reagents and Materials:
- Phosphate-Buffered Saline (PBS), sterile
- Collagenase Type I or Liberase (0.1% working concentration in PBS) [8]
- Complete growth medium (e.g., α-MEM or DMEM/F12 supplemented with 5-10% Human Platelet Lysate (HPL) or FBS) [56]
- Recombinant trypsin (e.g., TrypLE Select) for passaging
- Antibiotic/Antimycotic solution (for initial processing only)
- Cell strainers (100 μm)
- Tissue culture plasticware
Procedure:
- Tissue Collection and Processing: Obtain adipose tissue (lipoaspirate or solid tissue) under informed consent and approved ethical guidelines. Wash the tissue extensively with PBS containing antibiotics to reduce blood-derived contaminants and microbial load [12].
- Enzymatic Digestion: Mince the washed tissue into small fragments (< 1-4 mm³) and transfer to a digestion solution containing 0.1% Collagenase Type I or Liberase. Incubate at 37°C for 2-3 hours with constant agitation. Optimization Note: For bovine adipose tissue, 0.1% Liberase for 3 hours provided the highest cell yield with low population doubling time [8].
- Enzyme Neutralization and Fraction Separation: Add an equal volume of complete growth medium to neutralize the enzymatic activity. Centrifuge the digestate at a low speed (300-500 x g) for 5-10 minutes. This will separate the mixture into three layers: a top lipid layer, a middle aqueous layer, and a bottom SVF cell pellet.
- SVF Collection and Washing: Carefully aspirate the top and middle layers. Resuspend the SVF pellet in fresh complete medium and filter the suspension through a 100 μm cell strainer to remove any undigested tissue fragments [12].
- Cell Seeding and Culture: Seed the filtered cells into culture flasks at an appropriate density. Place the flasks in a 37°C incubator with 5% CO₂. Perform the first medium change after 48-72 hours to remove non-adherent cells, then change the medium every 2-3 days thereafter.
- Cell Passaging and Expansion: Once cells reach 85-95% confluence, harvest them using a recombinant trypsin solution (e.g., TrypLE Select) for subsequent passaging and expansion [56].
The Scientist's Toolkit: Essential Reagents for MSC Isolation
Successful MSC isolation and culture depend on a carefully selected set of reagents and tools. The table below details key materials and their specific functions in the process.
Table 3: Key Research Reagent Solutions for MSC Isolation
| Reagent / Material | Function in Protocol | Notes for Optimization |
|---|---|---|
| Collagenase Type I / Liberase [8] [12] | Enzymatically digests the extracellular matrix of adipose tissue to release the SVF. | Liberase can offer higher yield and viability compared to Collagenase I alone for some tissue types [8]. |
| Human Platelet Lysate (HPL) [56] | Serum-free, xeno-free supplement for cell culture medium; provides essential growth factors and adhesion factors. | Superior to FBS for clinical applications; can be used at 2-10% concentration. Supplementing serum-free media with 2% HPL can enhance primary culture output [56]. |
| Recombinant Trypsin (TrypLE) [56] | Enzyme for detaching adherent MSCs during passaging; animal-origin free. | Gentler on cells than porcine trypsin, supporting higher post-passage viability. |
| Cell Strainers (70μm, 100μm) [12] | Removes undigested tissue clumps and debris from the cell suspension post-digestion. | Critical for obtaining a single-cell suspension for counting and seeding. |
| Quality Control Instrumentation [54] [57] | Monitors cleanroom compliance and analyzes cell populations. | Technologies like AMIL link ensure aseptic conditions. Instruments like Celector use label-free fractionation for QC of cell isolates [54] [57]. |
Concluding Remarks and Future Perspectives
Addressing the challenges of low cell yield, poor viability, and microbial contamination in MSC isolation requires a multifaceted strategy grounded in rigorous protocol optimization and systematic quality control. The integration of advanced reagents like Liberase and HPL, coupled with precise process control and emerging quality assessment technologies, provides a robust pathway toward manufacturing MSCs that meet the stringent standards required for clinical translation. As the field progresses, the adoption of these optimized, standardized protocols will be paramount in ensuring the reproducible production of high-quality MSCs, thereby unlocking their full therapeutic potential across a widening spectrum of human diseases.
Within the broader scope of research on enzymatic digestion protocols for isolating Mesenchymal Stromal Cells (MSCs), the selection of an optimal dissociation enzyme is a critical determinant of yield, viability, and functionality. This application note provides a data-driven case study on the optimization of bovine adipose-derived MSC (BvAdMSC) isolation, with a specific focus on the use of Liberase. The protocol is particularly relevant for fields requiring high cell yields, such as cultured meat production and veterinary regenerative medicine [8] [58].
Comparative Enzyme Efficiency Data
A comprehensive study evaluated 32 distinct isolation conditions to identify the optimal protocol for isolating BvAdMSCs from subcutaneous adipose tissue. Key enzymatic mixtures, including Collagenase type I (Coll IA), Collagenase type I + Trypsin, Liberase (LibTM), and Collagenase type IV, were tested at varying concentrations and incubation times [8].
Table 1: Cell Yield and Proliferation of BvAdMSCs Isolated Under Different Enzymatic Conditions
| Enzyme Mixture | Concentration | Incubation Time | Average Cell Yield (x10⁶ cells/g tissue) | Population Doubling Time | Success Rate (Donors Responding) |
|---|---|---|---|---|---|
| Liberase | 0.1% | 3 hours | 30.48 - 67.10 | Low | High (≥5/8 donors) |
| Liberase | 0.1% | 6 hours | > 35.00 | Low | High (≥5/8 donors) |
| Liberase | 0.1% | Overnight | > 35.00 | Low | High (≥5/8 donors) |
| Collagenase Type I | 0.1% | 6 hours | > 35.00 | Low | High (≥5/8 donors) |
| Collagenase Type I | 0.04% | 6 hours | > 35.00 | Low | High (≥5/8 donors) |
| Collagenase Type IV | 0.04% | 6 hours | No plastic-adherent cells | N/A | 0/8 donors |
The data conclusively showed that isolation with 0.1% Liberase for 3 hours yielded the highest number of cells per gram of tissue while maintaining a low population doubling time, indicating robust proliferation potential. This condition was statistically superior to the commonly used Collagenase Type I at the same concentration and duration [8]. Furthermore, cell viability exceeded 95% across all successful isolation conditions [8].
Detailed Experimental Protocol for BvAdMSC Isolation with Liberase
Reagents and Materials
- Liberase (e.g., Sigma, Roche MTF C/T GMP grade) [8] [59]
- Collection Medium: Phosphate-buffered saline (PBS) supplemented with 1% penicillin-streptomycin (P/S) [58]
- Digestion Medium: Alpha Minimum Essential Medium (MEM α) supplemented with 0.1% (w/v) Liberase, 1% P/S, and 50 µg/mL nystatin [8] [58]
- Culture Maintenance Medium: MEM α supplemented with 15-30% Fetal Bovine Serum (FBS), 1% P/S, 50 µg/mL nystatin, and 80 µg/mL amikacin sulfate [8] [58] [60]
- Surgical tools (scalpel, scissors, forceps)
- 70 µm cell strainer
Step-by-Step Procedure
- Tissue Collection and Transport: Aseptically collect subcutaneous adipose tissue from the tailhead area of a cow [58]. Transport the tissue to the lab within 1-2 hours in cold PBS with antibiotics [60].
- Tissue Processing: Wash the tissue extensively with sterile cold saline containing high-dose antibiotics. Using sterile instruments, mince the tissue into small pieces of approximately 1 mm³ [58] [60].
- Enzymatic Digestion:
- Reaction Neutralization and Cell Harvest:
- Neutralize the enzymatic reaction by adding an equal volume of Culture Maintenance Medium containing FBS [8] [60].
- Filter the resulting cell suspension through a 70 µm cell strainer to remove debris and undigested tissue fragments [60].
- Centrifuge the filtrate at 300-400 × g for 5-10 minutes to pellet the cells [58] [60].
- Red Blood Cell (RBC) Lysis (Optional): Resuspend the cell pellet in an RBC lysis buffer for 10 minutes at room temperature to eliminate erythrocyte contamination. Centrifuge again to obtain the final Stromal Vascular Fraction (SVF) pellet [61].
- Primary Cell Culture: Resuspend the final cell pellet in Culture Maintenance Medium and seed in a culture flask. Incubate at 38.5°C with 5% CO₂ [60]. Replace the medium after 24 hours to remove non-adherent cells. Refresh the medium 2-3 times per week [8] [60].
Figure 1: Workflow for the isolation of Bovine Adipose-derived Mesenchymal Stromal Cells (BvAdMSCs) using the optimized Liberase protocol.
Post-Isolation Characterization and Myogenic Differentiation
Confirmation of MSC Identity
Cells isolated using the optimized protocol must be characterized to confirm their MSC identity, adhering to established bovine and international guidelines [58] [60].
- Plastic Adherence: Isolated BvAdMSCs should exhibit a spindle-shaped, fibroblast-like morphology and adhere to plastic surfaces under standard culture conditions [8] [58].
- Immunophenotyping: Flow cytometry analysis should confirm the expression of characteristic MSC surface markers CD73, CD90, CD105, and CD44, while lacking expression of the hematopoietic marker CD45 [58] [60]. Note that subpopulations of bovine AD-MSCs may express CD34 [60].
- Tri-lineage Differentiation: The cells must demonstrate the capacity to differentiate into adipocytes, osteocytes, and chondrocytes under specific inductive conditions in vitro [8] [58].
Myogenic Differentiation for Cultured Meat Applications
For cultured meat production, BvAdMSCs can be directed toward the myogenic lineage.
- Induction Protocol: Treat confluent BvAdMSCs with 5-aza-2'-deoxycytidine and galectin-1 to induce myogenic differentiation [8].
- Molecular Validation: Differentiated cells show upregulated mRNA levels of key myogenic regulatory factors (MRFs), including MYF5, MYOD1, MYF6, and myogenin (MYOG), while PAX3 expression decreases [8].
- Protein Confirmation: Immunofluorescence staining confirms the presence of skeletal muscle proteins such as desmin (DES), tropomyosin (TM), and myosin heavy chain (MyHC) [8].
The Scientist's Toolkit: Key Research Reagent Solutions
Table 2: Essential Reagents for BvAdMSC Isolation and Culture
| Reagent | Function in Protocol | Key Considerations |
|---|---|---|
| Liberase | Purified enzyme blend (Collagenase I/II & Thermolysin) for tissue dissociation. | Superior to Collagenase I for bovine adipose; ensure mammalian tissue-free (MTF) grade for safety [8] [59]. |
| Fetal Bovine Serum (FBS) | Component of culture medium for cell growth and enzyme neutralization. | High concentration (15-30%) is often used for primary bovine MSC culture [8] [60]. |
| Antibiotic-Antimycotic | Prevents microbial contamination in tissue and cell cultures. | Crucial for primary tissue isolation; can include penicillin/streptomycin and nystatin [58]. |
| Cell Strainers (70-100 µm) | Removes undigested tissue fragments and debris from the cell suspension. | Essential for obtaining a clean stromal vascular fraction (SVF) [60] [61]. |
| Trypsin-EDTA | Passaging and sub-culturing of adherent BvAdMSCs. | Standard reagent for detaching adherent cells [60]. |
Impact of Donor Characteristics
Research indicates that donor characteristics significantly influence the properties of isolated MSCs. Studies on bovine AD-MSCs show that donor age and breed can affect proliferation capacity, differentiation potential, and immunophenotype [60].
- Age: Fetal and calf-derived MSCs often demonstrate a higher proliferation capacity compared to those from adult animals [60].
- Breed: Significant differences in osteogenic differentiation potential and surface marker expression (e.g., CD34) have been observed between breeds like Holstein Friesian and Belgian Blue [60]. These factors should be considered when designing experiments or production processes where consistent MSC performance is critical.
The enzymatic digestion protocol used to isolate Mesenchymal Stromal Cells (MSCs) from their native tissue is a critical determinant of the resulting cell population's fundamental biological characteristics and therapeutic potential. This initial step influences not only immediate cell yield and viability but also long-term culture performance, including proliferation capacity, senescence rates, and the composition of the secreted factor profile (secretome) [8] [62]. As MSC-based therapies advance toward clinical translation, achieving batch-to-batch consistency requires standardized, optimized digestion methods that maximize cell yield without compromising function [49]. This Application Note synthesizes recent research to provide detailed, quantitative guidelines on how digestion parameters impact MSC biology and to present standardized protocols for isolating functionally superior MSCs.
Quantitative Impact of Digestion on MSC Attributes
The choice of enzyme, its concentration, and the duration of incubation collectively dictate the efficiency of MSC isolation and the quality of the harvested cells. Systematic evaluations of these parameters are essential for protocol optimization.
Table 1: Impact of Enzymatic Digestion on MSC Yield and Proliferation
| Enzyme / Mixture | Concentration | Incubation Time | Average Cell Yield (cells/g tissue) | Impact on Proliferation / Viability |
|---|---|---|---|---|
| Liberase [8] | 0.1% | 3 hours | 67.1 × 10⁶ | Highest yield; low population doubling time; viability >95% |
| Liberase [8] | 0.1% | 6 hours | 48.75 × 10⁶ | Viability >95% |
| Collagenase Type I + Trypsin [8] | 0.1% | 3 hours | 35.98 × 10⁶ | Viability >95% |
| Collagenase Type I [8] | 0.1% | 6 hours | ~35 × 10⁶ | Viability >95% |
| Collagenase Type I [8] | 0.04% | 6 hours | ~35 × 10⁶ | >5 CFU reached in 5 days |
| Vibrio alginolyticus Collagenase [62] | 3.6 mg/mL | 20 minutes | Comparable to standard methods | High viability and clonogenic capacity; reduced stress on expanded cells |
| Clostridium histolyticum Collagenase [62] | Standard | 45 minutes | Comparable to above | Can reduce vitality of expanded cells |
Table 2: Impact of Digestion on Long-Term MSC Function and Secretome
| Isolation/Culture Method | Impact on Senescence & Apoptosis | Impact on Secretome & Potency | Key Functional Outcomes |
|---|---|---|---|
| 3D Bio-Block Culture (Post-Expansion) [63] | Senescence reduced by 30-37%; Apoptosis decreased 2-3-fold | Secretome protein content preserved; EV production increased ~44% | Enhanced proliferation; higher trilineage differentiation; robust regenerative potency in vivo |
| 3D Spheroid Culture (Post-Expansion) [63] | Not specified | EV production declined 30-70%; Secretome protein declined 47% | Induced senescence and apoptosis in target endothelial cells |
| Microcarrier-Microbioreactor Culture [64] | Improved batch-to-batch reproducibility | Enhanced expression of key therapeutic genes (e.g., IDO1, IL-10) for ARDS | Reduced phenotypic heterogeneity; primed secretome for specific clinical indication |
| Vibrio alginolyticus Collagenase [62] | Not specified | Not specified | Protected vital structures for tissue restructuration; high selectivity for collagen |
Detailed Experimental Protocols
Optimized Protocol for Bovine Adipose Tissue Digestion
This protocol, adapted from a study on bovine adipose-derived MSCs, is designed to maximize cell yield and function [8].
- Tissue Collection and Preparation: Obtain subcutaneous adipose tissue aseptically. Rinse the tissue thoroughly with sterile PBS supplemented with 1-2% antibiotics (e.g., Penicillin-Streptomycin) to remove blood residues. Mince the tissue into fine pieces (< 1 cm³) using sterile scalpels or scissors.
- Enzymatic Digestion: Suspend the minced tissue in a solution of 0.1% Liberase in a suitable buffer (e.g., PBS). Use a volume of digestion solution that is 3-5 times the volume of the tissue.
- Incubation: Incubate the tissue suspension for 3 hours at 37°C with constant agitation (e.g., on an orbital shaker or with periodic manual shaking) to ensure uniform digestion.
- Digestion Neutralization and Filtration: After incubation, neutralize the enzyme by adding an equal volume of complete culture medium (e.g., α-MEM or DMEM supplemented with serum or human platelet lysate). Mix thoroughly. Filter the resulting cell suspension through a 100 µm cell strainer to remove undigested tissue fragments and debris.
- Cell Recovery: Centrifuge the filtered suspension at 300-400 × g for 10 minutes. Carefully aspirate the supernatant, including the floating adipocytes and oil droplets. Resuspend the cell pellet in fresh complete culture medium.
- Cell Seeding and Culture: Seed the cells in culture flasks at a density of 0.5-1.0 × 10⁵ cells/cm². Place the flasks in a 37°C incubator with 5% CO₂. Perform the first medium change after 48-72 hours to remove non-adherent cells, and subsequently change the medium every 2-3 days.
Rapid Protocol Using Novel Collagenase for Human Adipose Tissue
This protocol utilizes a novel enzyme for a faster, gentler isolation process suitable for clinical applications [62].
- Tissue Collection: Collect lipoaspirate (e.g., from abdominal area) using standard surgical procedures under informed consent. Transport in an adiabatic container and process within 24 hours.
- Enzymatic Digestion: Add 3.6 mg/mL of Vibrio alginolyticus-based collagenase to the lipoaspirate sample.
- Incubation: Incubate the mixture for 20 minutes at 37°C.
- Process Termination and Washing: Stop the reaction by adding cold complete medium. Centrifuge the suspension to pellet the cells.
- Cell Seeding and Expansion: Seed the resulting Stromal Vascular Fraction (SVF) cells in culture flasks. Expand the cells and confirm MSC phenotype through surface marker expression (CD73+, CD90+, CD105+, CD34-, CD45-) and trilineage differentiation potential.
Signaling Pathways and Experimental Workflows
The following diagrams illustrate the logical relationship between digestion parameters and MSC functional outcomes, as well as a general workflow for assessing digestion impact.
Diagram 1: Digestion Parameters Influence on MSC Function
Diagram 2: Workflow for MSC Digestion Impact Analysis
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Key Reagents for Optimized MSC Isolation and Culture
| Reagent / Material | Function / Role | Example Use Case / Note |
|---|---|---|
| Liberase [8] | Enzyme blend for tissue dissociation. | High-purity GMP-grade enzyme; optimal for adipose tissue at 0.1% for 3h. |
| Vibrio alginolyticus Collagenase [62] | Highly selective bacterial collagenase. | Protects cell surface proteins; enables shorter (20 min) digestion. |
| Clostridium histolyticum Collagenase [8] [62] | Conventional bacterial collagenase. | Common but less selective; can reduce vitality of expanded cells. |
| Human Platelet Lysate (HPL) [56] | Serum-free culture supplement. | Xeno-free FBS alternative; supports clinical-scale expansion. |
| Recombinant Trypsin (TrypLE) [56] | Animal-origin-free enzyme for passaging. | Gentle cell detachment; improves process consistency. |
| Dissolvable Gelatin Microcarriers [64] | 3D substrate for scalable expansion. | Enhances secretome and reduces senescence vs. 2D culture. |
| Serum-Free Media (e.g., NutriStem XF) [56] | Defined, xeno-free basal medium. | Improves cell uniformity and supports specific secretory profiles. |
The enzymatic digestion protocol is a foundational step that profoundly influences the yield, phenotypic stability, and functional potency of isolated MSCs. Data demonstrates that optimized protocols using specific enzymes like Liberase or novel alternatives such as Vibrio alginolyticus collagenase can significantly enhance cell yield, reduce population doubling time, and improve the quality of the resulting cell product [8] [62]. Furthermore, post-isolation culture environments, including 3D systems like Bio-Blocks and controlled bioreactors, can mitigate long-term functional decline by reducing senescence and preserving a therapeutically relevant secretome [63] [64]. Adhering to the detailed protocols and reagent solutions provided here will enable researchers to standardize the production of high-quality MSCs, thereby enhancing the reliability and efficacy of pre-clinical and clinical studies.
The transition of Mesenchymal Stromal Cell (MSC) therapies from laboratory research to commercial therapeutics necessitates the development of robust, scalable, and automated production processes. Enzymatic digestion protocols for MSC isolation form the critical foundation upon which industrial-scale biomanufacturing depends. Current research demonstrates significant progress in standardizing these isolation methods across different tissue sources, particularly adipose tissue and umbilical cord, which offer abundant MSC yields suitable for large-scale production [28]. The optimization of enzymatic parameters—including enzyme selection, concentration, digestion time, and temperature—has become paramount for maximizing cell yield while maintaining viability, proliferative capacity, and therapeutic functionality [8] [30]. This application note provides a comprehensive analysis of current enzymatic digestion methodologies, quantitative comparisons of isolation efficiency, detailed protocols for different tissue sources, and a strategic framework for implementing these processes in automated bioreactor systems for industrial-scale MSC manufacturing.
Quantitative Analysis of Enzymatic Digestion Efficiency
Comparative Evaluation of Enzymes and Parameters
Table 1: Enzymatic Digestion Efficiency for Adipose-Derived MSC Isolation
| Enzyme/Combination | Concentration | Incubation Time | Average Cell Yield (×10^6 cells/g tissue) | Cell Viability (%) | Time to First Passage (days) |
|---|---|---|---|---|---|
| Liberase | 0.1% | 3 hours | 30.48-67.10 | >95% | 5 |
| Collagenase Type I | 0.1% | 3 hours | 21.50-45.20 | >95% | 5-7 |
| Collagenase Type I | 0.1% | 6 hours | 25.10-52.30 | >95% | 5-7 |
| Collagenase Type I + Trypsin | 0.1% | 3 hours | 18.60-40.80 | >95% | 6-7 |
| Collagenase Type IV | 0.1% | 3 hours | 15.20-35.10 | >95% | 6-8 |
| Collagenase Type I | 0.04% | 6 hours | 22.10-48.90 | >95% | 5 |
| Liberase | 0.1% | 6 hours | 35.20-65.80 | >95% | 5 |
| Liberase | 0.1% | Overnight | 28.90-62.10 | >95% | 5-6 |
Table 2: Enzymatic Digestion Efficiency for Umbilical Cord-Derived MSC Isolation
| Parameter | Optimal Condition | Alternative Conditions Tested | Impact on P0 Cell Yield |
|---|---|---|---|
| Collagenase NB6 Concentration | 0.4 PZ U/mL | 0.2, 0.6 PZ U/mL | Highest yield at 0.4 PZ U/mL |
| Digestion Time | 3 hours | 2, 4 hours | Direct correlation up to 3 hours |
| Seeding Density | 1g tissue/75cm² flask | 0.5g, 2g tissue/75cm² flask | Density-dependent yield |
| Culture Medium | 2-5% hPL | 10% hPL, Serum-free variants | Comparable results with 2-5% hPL |
Critical Analysis of Isolation Efficiency Data
The systematic evaluation of 32 isolation conditions for bovine adipose-derived MSCs revealed that Liberase at 0.1% concentration for 3 hours provided the highest cell yield while maintaining excellent viability and proliferation potential [8]. This optimized protocol achieved average yields of 30.48-67.10 × 10^6 cells/g tissue, significantly outperforming standard Collagenase Type I protocols under equivalent conditions. Similarly, for human umbilical cord-derived MSCs, optimization studies identified 0.4 PZ U/mL Collagenase NB6 with a 3-hour digestion as the optimal parameters for maximizing primary cell yield while maintaining GMP compliance [30].
Notably, increasing enzyme concentration beyond optimal levels or extending digestion times did not consistently improve yields and potentially risks compromising cell viability and functionality [8]. The data demonstrates a positive correlation between umbilical cord tissue weight and P0 WJ-MSC yield, highlighting the importance of standardized tissue collection and processing methods for predictable scaling [30]. These quantitative comparisons provide critical baseline data for designing scaled-up isolation processes in bioreactor systems.
Detailed Experimental Protocols for MSC Isolation
Adipose-Derived MSC Isolation Protocol
Materials Required:
- Lipoaspirate or solid adipose tissue (abdominal region preferred)
- Sterile phosphate-buffered saline (PBS)
- Liberase (0.1% working concentration) or Collagenase Type I (0.1% working concentration)
- Complete growth medium: αMEM supplemented with 2mM L-glutamine, 100 IU/mL penicillin, 0.1 mg/mL streptomycin, and 10-20% FBS or human platelet lysate
- Cell strainers (100μm and 40μm)
- Centrifuge tubes (50mL)
- Water bath or incubator with agitation capability
Step-by-Step Procedure:
Tissue Preparation: Wash adipose tissue samples extensively with PBS to remove blood contaminants and connective tissue. For lipoaspirate, transfer to 50mL centrifuge tubes and allow fat to separate from infranatant by gravity or low-speed centrifugation (300g for 5 minutes) [12].
Enzymatic Digestion: Add pre-warmed enzyme solution (0.1% Liberase or Collagenase Type I) to tissue at a ratio of 1:1-1:3 (v/v, enzyme:tissue). Incubate at 37°C with continuous agitation for 3 hours for Liberase or 3-6 hours for Collagenase Type I [8].
Digestion Termination: Neutralize enzyme activity by adding complete growth medium containing serum at a 1:1 ratio. Mix thoroughly by inversion.
Stromal Vascular Fraction Separation: Centrifuge the digest at 1200g for 10 minutes to separate the stromal vascular fraction (SVF) pellet from adipocytes and debris [12] [43].
Cell Filtration and Seeding: Resuspend the SVF pellet in complete growth medium and filter sequentially through 100μm and 40μm cell strainers to remove tissue fragments. Seed cells at a density of 1×10^3 cells/cm² in culture vessels [12].
Culture and Expansion: Incubate cultures at 37°C in 5% CO₂ with medium changes every 2-3 days. Passage cells at 80% confluence using standard trypsinization protocols [43].
Umbilical Cord-derived MSC Isolation Protocol
Materials Required:
- Human umbilical cord tissue (>20cm length, processed within 24-48 hours post-collection)
- GMP-grade Collagenase NB6 (0.4 PZ U/mL working concentration)
- Decontamination solution: 0.5% povidone-iodine
- DPBS without calcium or magnesium
- Serum-free culture medium (e.g., NutriStem) supplemented with 2-5% human platelet lysate
- Surgical scalpels and forceps
Step-by-Step Procedure:
Cord Processing: Rinse umbilical cord with DPBS to remove blood contaminants. Decontaminate with 0.5% povidone-iodine solution for 3 minutes, followed by three DPBS rinses [30].
Vessel Removal and Tissue Mincing: Dissect cord to expose Wharton's jelly and carefully remove two arteries and one vein. Mince the remaining Wharton's jelly tissue into 1-4mm³ fragments using surgical scalpels [30] [27].
Enzymatic Digestion: Transfer tissue fragments to enzyme solution (0.4 PZ U/mL Collagenase NB6) at a ratio of 1g tissue per 5-10mL enzyme. Incubate at 37°C for 3 hours with intermittent agitation [30].
Cell Collection and Seeding: Neutralize digestion with complete medium and filter through 100μm cell strainer. Centrifuge at 1200g for 10 minutes and resuspend pellet in culture medium. Seed at optimal density of 1g original tissue per 75cm² flask [30].
Culture Maintenance: Incubate at 37°C in 5% CO₂ with medium changes twice weekly. Passage cells at 80-90% confluence using standard detachment protocols [65] [27].
Workflow Integration for Automated Bioreactor Systems
Figure 1: Integrated Workflow for Automated MSC Biomanufacturing
The transition to automated bioreactor systems requires careful integration of optimized isolation protocols with scalable expansion technologies. Current research indicates that passages 2-5 demonstrate the highest viability and proliferation capacity, making these generations ideal for bioreactor seeding [30]. The enzymatic isolation process serves as the critical initial step that determines both the quantity and quality of cells entering the expansion system. Implementation of process analytical technologies (PAT) for real-time monitoring of critical quality attributes during both isolation and expansion phases enables automated feedback control for consistent product quality [30] [27].
The Scientist's Toolkit: Essential Reagent Solutions
Table 3: Key Research Reagent Solutions for MSC Isolation and Expansion
| Reagent Category | Specific Products/Functions | Application Notes | Industrial Scale Considerations |
|---|---|---|---|
| Digestion Enzymes | Liberase, Collagenase Type I, Collagenase NB6 GMP | Liberase optimal for adipose tissue; Collagenase NB6 for GMP-compliant UC processing | GMP-grade essential for clinical applications; enzyme consistency critical for batch-to-batch reproducibility |
| Culture Media | αMEM, DMEM, Serum-free formulations (e.g., NutriStem) | αMEM superior for UC-MSC expansion; DMEM common for AD-MSC | Transition from FBS to hPL or defined serum-free formulations for clinical compliance |
| Supplementation | Fetal Bovine Serum (FBS), Human Platelet Lysate (hPL) | 2-5% hPL provides comparable expansion to 10% FBS | hPL reduces xenogenic components; requires rigorous quality control and pooling strategies |
| Dissociation Agents | Trypsin-EDTA, TrypLE Select | Standard trypsinization for passaging; TrypLE as animal-origin-free alternative | Enzyme-free dissociation methods preferable for automated systems to minimize process variability |
| Cryopreservation | DMSO, Defined cryopreservation media | Standard 10% DMSO protocol; clinical-grade cryomedium alternatives | Controlled-rate freezing critical; final container configuration dependent on administration route |
Strategic Implementation for Industrial Translation
Process Optimization and Scalability
Successful transition to industrial-scale bioreactors requires meticulous attention to parameter optimization at the isolation stage. Research demonstrates that enzymatic digestion efficiency directly impacts downstream expansion capability in bioreactor systems [8] [30]. For adipose-derived MSCs, the optimized Liberase protocol reduces initial processing time while maximizing cell yield, providing more viable cells for bioreactor inoculation. For umbilical cord-derived MSCs, the standardized enzymatic approach with Collagenase NB6 ensures consistent isolation efficiency across multiple donors, addressing a critical challenge in allogeneic therapy manufacturing [30] [27].
The integration of automated closed-system processing for the enzymatic digestion phase enables seamless transfer to expansion bioreactors while maintaining sterility and process control. Recent advances demonstrate that mechanical isolation methods, while faster and more cost-effective, yield comparable cell viability and proliferation characteristics to enzymatic methods, presenting an alternative approach for certain applications [6]. However, enzymatic methods currently provide superior consistency and characterization capabilities for regulated therapeutic applications.
Quality by Design Framework
Implementing a Quality by Design (QbD) approach to enzymatic isolation parameters establishes the foundation for robust industrial processes. Critical process parameters (CPPs) including enzyme concentration, digestion time, temperature, and agitation rate must be systematically evaluated for their impact on critical quality attributes (CQAs) such as cell viability, identity, potency, and differentiation capacity [8] [30]. The quantitative data presented in this application note provides initial design space parameters for these optimization activities.
Donor-to-donor variability represents a significant challenge in MSC biomanufacturing. Recent studies involving 90 umbilical cord donors demonstrate that robust enzymatic processing methods can minimize technical variability, thereby enhancing product consistency despite biological differences [27]. This standardization is essential for obtaining regulatory approval and ensuring reproducible therapeutic effects.
The scalability and automation of MSC manufacturing processes depend fundamentally on robust, optimized enzymatic isolation protocols that can be seamlessly integrated with bioreactor expansion technologies. The quantitative data and detailed methodologies presented in this application note provide a scientific foundation for designing and implementing industrial-scale MSC production systems. By adopting standardized enzymatic digestion parameters, implementing QbD principles, and utilizing the essential reagent solutions outlined, researchers and manufacturers can accelerate the translation of MSC therapies from research laboratories to clinical applications. Continued refinement of these processes, particularly through the integration of advanced process analytics and automation technologies, will further enhance the efficiency, consistency, and scalability of MSC manufacturing for therapeutic applications.
Ensuring Quality: Validation, Characterization, and Method Comparison
International Society for Cell Therapy (ISCT) Standards for MSC Validation
The International Society for Cell & Gene Therapy (ISCT) has established critical updates to the identification and validation standards for Mesenchymal Stromal Cells (MSCs), marking a significant evolution from the 2006 position statement [66]. These changes respond to nearly two decades of scientific debate and advancement, refining the fundamental understanding of MSC biology and establishing a more rigorous framework for their therapeutic development [66] [3]. The most striking clarification involves the formal definition of "MSC" as Mesenchymal Stromal Cells rather than "Mesenchymal Stem Cells," unless researchers provide experimental evidence of true stem cell properties such as self-renewal and multi-lineage differentiation potential [66]. This terminological precision addresses longstanding reproducibility issues and reflects a more scientifically accurate understanding of these cells' primary mechanisms of action, which often involve paracrine signaling rather than true stem cell differentiation [3].
Table 1: Comparison of ISCT 2006 vs. 2025 MSC Identification Standards
| Standard Element | 2006 Standard | 2025 Standard |
|---|---|---|
| Cell Definition | Mesenchymal Stem Cells (MSCs) | Mesenchymal Stromal Cells (MSCs) |
| Stemness Requirement | Must demonstrate trilineage differentiation | Must provide evidence to use the term "stem" |
| Marker Detection | Qualitative (positive/negative) | Quantitative (thresholds and percentages) |
| Tissue Origin | Not emphasized | Must be specified and considered |
| Critical Quality Attributes | Not required | Must assess efficacy and functional properties |
| Culture Conditions | No standard reporting requirement | Detailed parameter reporting required |
The updated standards emerged from dedicated ISCT workshops where key opinion leaders in MSC biology, clinical application, and regulatory science gathered to address the pressing need for standardization in clinical trial design, conduct, and reporting [9]. This collective expertise has yielded a more transparent, reproducible pathway for MSC therapeutic development, ensuring that meta-analyses can be generated from comparable datasets and that these advanced therapeutic medicinal products can successfully transition to market [9].
Updated ISCT Validation Criteria and Critical Quality Attributes
The 2025 ISCT standards introduce comprehensive upgrades to MSC identification criteria, moving beyond minimal definitions to establish robust characterization frameworks essential for clinical applications [66].
Phenotypic Marker Specifications
The updated standards maintain CD73, CD90, and CD105 as fundamental positive markers but introduce stricter quantitative requirements [66]. Researchers must now specify the precise threshold percentage for positive identification via flow cytometry, moving from qualitative assessment to quantitative reporting. Furthermore, the inclusion of the hematopoietic marker CD45 as a mandatory negative marker is emphasized to ensure population purity. Crucially, the standards now require complete reporting of results for each marker, including the exact percentage of positive cells, to enhance data transparency and comparability across studies and laboratories [66].
Functional and Efficacy Characterization
A paradigm shift in the 2025 standards is the incorporation of efficacy and functional characterization into Critical Quality Attributes (CQAs) [66]. This evolution acknowledges that MSC products must not only meet phenotypic standards but also demonstrate predicted clinical functionality. The standards emphasize that functional validation assays should be specifically designed around the intended clinical application, ensuring that the measured attributes directly relate to therapeutic mechanism of action [9]. For autoimmune disease applications, for instance, this might include validated immunomodulatory potency assays rather than relying solely on traditional differentiation potential [9].
The standards also place new emphasis on specifying the tissue origin of MSCs, recognizing that cells from different anatomical sources may exhibit distinct phenotypic and functional properties despite sharing core markers [66]. This tissue-specific perspective aligns with advances in single-cell omics technologies that have revealed previously unappreciated heterogeneity in stromal cell populations from various sources.
Isolation Techniques and Enzymatic Digestion Protocols
The selection of an appropriate isolation method represents a critical initial step in obtaining a high-quality MSC population suitable for therapeutic applications. The enzymatic digestion method offers distinct advantages for clinical-scale manufacturing, including higher cell yields in shorter timeframes and superior reproducibility [67] [8].
Enzymatic Digestion Workflow
The following diagram illustrates the generalized workflow for enzymatic isolation of MSCs from solid tissues:
Comparative Analysis of Enzymatic Conditions
Research systematically evaluating enzymatic protocols provides crucial quantitative data for optimizing isolation efficiency. A comprehensive 2024 study compared 32 different isolation conditions for bovine adipose tissue-derived MSCs, testing four enzyme mixtures at varying concentrations and incubation times [8]. The findings demonstrate that protocol optimization can significantly impact cell yield and quality.
Table 2: Evaluation of Enzymatic Protocols for Adipose Tissue-Derived MSC Isolation
| Enzyme Mixture | Concentration | Incubation Time | Average Cell Yield (×10^6 cells/g tissue) | Time to First Passage (days) |
|---|---|---|---|---|
| Liberase | 0.1% | 3 hours | 67.1 | 5 |
| Collagenase IA | 0.1% | 3 hours | 35.2 | 7 |
| Collagenase IA + Trypsin | 0.1% | 3 hours | 41.5 | 6 |
| Collagenase IV | 0.1% | 3 hours | 15.8 | 9 |
| Liberase | 0.1% | 6 hours | 58.3 | 5 |
| Liberase | 0.1% | Overnight | 46.7 | 6 |
The data revealed that Liberase at 0.1% concentration for 3 hours provided the highest cell yield combined with the shortest time to first passage [8]. This optimized protocol yielded approximately 90% more cells than the commonly used Collagenase IA under the same conditions, demonstrating the significant impact of enzyme selection on isolation efficiency. Importantly, cells isolated using this optimized protocol maintained tri-lineage differentiation potential and expressed characteristic MSC surface markers, confirming that isolation efficiency was not achieved at the expense of cell quality [8].
Tissue-Specific Digestion Considerations
Different tissue sources require modifications to the general enzymatic digestion protocol to account for variations in extracellular matrix composition and tissue density [67]:
- Adipose Tissue: Digest with 0.8 mg/ml collagenase for 4 hours at 37°C with continuous shaking [67]
- Tendon Tissue: Requires more extensive digestion with 5.6 mg/ml collagenase for 6 hours due to dense collagenous structure [67]
- Umbilical Cord Matrix: Digest with 2.4 mg/ml collagenase for 6 hours to liberate Wharton's jelly-derived MSCs [67]
Following digestion, the liberated mononuclear cell fraction is filtered (70μm pore size), centrifuged (437g for 5 minutes at 4°C), washed with PBS, and plated at densities ranging from 20,000-100,000 cells/cm² in appropriate culture medium [67].
The Scientist's Toolkit: Essential Reagents and Materials
The successful isolation and validation of MSCs according to ISCT standards requires carefully selected reagents and materials that comply with Good Manufacturing Practice (GMP) requirements where clinical application is intended [68].
Table 3: Research Reagent Solutions for MSC Isolation and Culture
| Reagent Category | Specific Examples | Function & Importance |
|---|---|---|
| Digestion Enzymes | Collagenase Type I, Liberase, Trypsin | Dissociates extracellular matrix to release mononuclear cells; critical for yield and viability [8] [67] |
| Basal Media | α-MEM, DMEM | Provides essential nutrients, vitamins, and buffers; choice affects proliferation and metabolism [68] |
| Serum Supplements | Fetal Bovine Serum (FBS), Human Platelet Lysate | Supplies growth factors and adhesion proteins; FBS raises xenogenic risk concerns [68] |
| Culture Surfaces | Tissue culture-treated plastic, GMP-compliant multilayer flasks | Provides adhesion substrate for selective MSC expansion; surface area scales with manufacturing needs [68] |
| Characterization Reagents | CD73, CD90, CD105, CD45 antibodies, differentiation induction kits | Confirms MSC identity per ISCT standards; essential for release criteria [66] [3] |
Experimental Protocol: MSC Isolation via Enzymatic Digestion
Materials and Pre-processing
Begin with tissue acquisition from approved sources (bone marrow aspirate, lipoaspirate, umbilical cord) with appropriate ethical oversight [68] [3]. For adipose tissue, collect specimens via tumescent liposuction without ultrasound assistance, as ultrasound has been shown to compromise MSC recovery and expansion capacity [68]. Process tissues within 24 hours of collection, maintaining sterile conditions throughout. Dissect away blood vessels and connective tissues, then mince the remaining tissue into 0.1-0.2 cm³ pieces using sterile surgical blades. Wash tissue pieces extensively with Hank's Balanced Salt Solution (HBSS) or phosphate-buffered saline (PBS) containing antibiotics (e.g., 0.1% gentamicin, 2.5 μg/ml amphotericin B) to reduce microbial contamination [67].
Enzymatic Digestion Procedure
Prepare digestion enzyme solution in HBSS at the optimal concentration determined for the specific tissue type (e.g., 0.1% Liberase for adipose tissue) [8]. Use approximately 3-5 ml of digestion solution per gram of tissue. Transfer the minced tissue to the enzyme solution and incubate in a continuously shaking water bath at 37°C for the appropriate duration:
Following digestion, filter the cell suspension through a 70μm cell strainer to remove undigested tissue fragments. Centrifuge the filtrate at 437g for 5 minutes at 4°C to pellet the stromal vascular fraction (for adipose tissue) or mononuclear cells (for other tissues) [67]. Resuspend the cell pellet in complete culture medium (e.g., DMEM supplemented with 20% FBS or clinical-grade human platelet lysate and antibiotics) [68].
Primary Culture and Expansion
Plate the resuspended cells at a density of 20,000-100,000 cells/cm² in tissue culture-treated flasks or plates. Maintain cultures in a humidified incubator at 37°C with 5% CO₂. Change the culture medium twice weekly to remove non-adherent cells and replenish nutrients [67]. Monitor daily for MSC adherence and colony formation. Passage cells when colonies reach 70-80% confluency using trypsin/EDTA solution. For clinical applications, document population doubling levels and establish a maximum passage number to prevent replicative senescence [68].
The following diagram illustrates the complete validation workflow following isolation:
The updated ISCT standards for MSC validation represent a critical advancement in the field, transitioning from minimal defining criteria to comprehensive characterization frameworks that better reflect the complexity of these therapeutic products [66]. The emphasis on quantitative rather than qualitative assessment, along with the new requirements for reporting tissue origin and culture conditions, addresses previously overlooked sources of variability in MSC research [9] [66].
For researchers developing enzymatic digestion protocols, the provided comparative data on enzyme efficiency offers evidence-based guidance for protocol optimization [8] [67]. The integration of these isolation methods with the updated validation criteria creates a standardized pathway from tissue acquisition to characterized cell product. This systematic approach enhances reproducibility across laboratories, facilitates more meaningful meta-analyses of clinical trial data, and ultimately supports the successful translation of MSC therapies from bench to bedside [9].
By implementing these comprehensive standards, the field moves closer to realizing the full therapeutic potential of mesenchymal stromal cells while maintaining the rigorous quality control necessary for advanced therapeutic medicinal products [9] [66].
Within the framework of research on enzymatic digestion protocols for the isolation of Mesenchymal Stromal Cells (MSCs), the definitive characterization of the resulting cell populations is paramount. Adherence to established international standards is not merely a procedural formality but a critical step to ensure the identity, purity, and functional potency of isolated cells, thereby guaranteeing the validity and reproducibility of research outcomes [3]. The International Society for Cell & Gene Therapy (ISCT) has defined minimal criteria for defining human MSCs, which include: (1) adherence to plastic under standard culture conditions; (2) specific surface antigen expression profile; and (3) tri-lineage differentiation potential into adipocytes, chondrocytes, and osteocytes [3] [25] [69]. This application note provides detailed protocols and analytical methods for the two cornerstone techniques fulfilling the latter two criteria: flow cytometry for immunophenotyping and trilineage differentiation for functional potency assessment.
Flow Cytometry for Immunophenotyping
Flow cytometry provides a quantitative analysis of the cell surface markers that uniquely identify MSCs and confirm the absence of contaminating hematopoietic cells [3] [69]. This technique allows for the simultaneous assessment of multiple markers on individual cells, providing data on the percentage of cells within a population that express the desired antigens.
Experimental Protocol
The following protocol is adapted for MSCs isolated from various tissues via enzymatic digestion [25] [8].
- Cell Preparation: Harvest MSCs at the 4th-6th passage upon reaching 80-90% confluence. Use trypsin-EDTA for cell detachment (incubate for 5 minutes at 37°C) [25].
- Washing: Neutralize the trypsin with complete culture medium, pellet the cells by centrifugation (e.g., 300-500 g for 5 minutes), and resuspend in flow cytometry buffer (e.g., DPBS containing 1-2% FBS).
- Staining: Aliquot approximately 1x10^5 to 1x10^6 cells per tube. Incubate cells with fluorochrome-conjugated antibodies against target antigens (see Table 1 for panel) for 20-30 minutes at 4°C in the dark. Include appropriate isotype controls for background subtraction.
- Washing and Fixation: Wash cells twice with flow cytometry buffer to remove unbound antibody. The cells can be analyzed immediately in buffer or fixed in a 1-4% paraformaldehyde solution if analysis is delayed.
- Data Acquisition and Analysis: Analyze the stained cell suspension using a flow cytometer. Collect a minimum of 10,000 events per sample. Use the isotype controls to set positive/negative gates. A population is considered positive if ≥95% of cells express the positive markers and ≤2% express the negative markers [69].
Table 1: Essential Surface Marker Panel for Human MSC Characterization via Flow Cytometry
| Marker Category | Marker | Acceptance Criterion (Positive Expression) | Function / Significance |
|---|---|---|---|
| Positive Markers | CD73 | ≥ 95% | Ecto-5'-nucleotidase; involved in purinergic signaling. |
| CD90 | ≥ 95% | Thy-1 cell adhesion molecule; a classic MSC marker. | |
| CD105 | ≥ 95% | Endoglin; part of the TGF-beta receptor complex. | |
| Negative Markers | CD11b/CD14 | ≤ 2% | Myeloid and monocyte lineage markers. |
| CD19/CD79α | ≤ 2% | B-cell lineage markers. | |
| CD34 | ≤ 2% | Hematopoietic progenitor and endothelial cell marker. | |
| CD45 | ≤ 2% | Pan-leukocyte marker. | |
| HLA-DR | ≤ 2% | MHC Class II antigen (unless immunologically activated). |
Trilineage Differentiation Assay
The trilineage differentiation assay is a functional test that confirms the multipotency of MSCs by inducing them to differentiate into adipocytes, osteocytes, and chondrocytes in vitro [25] [69]. The success of differentiation is typically confirmed by histological staining for lineage-specific markers and can be quantified using digital image analysis [69].
Experimental Protocol
The following protocol for 2D trilineage differentiation is based on established methods [25] [69]. Cells are seeded in specific densities and cultured in lineage-specific induction media for 2-4 weeks, with media changes every 3-4 days.
Table 2: Overview of Trilineage Differentiation Culture Conditions
| Lineage | Seeding Density | Key Induction Factors | Differentiation Time | Histological Stain |
|---|---|---|---|---|
| Adipogenesis | 3-5 x 10³ cells/cm² | Dexamethasone, IBMX, Indomethacin, Insulin | 14-21 days | Oil Red O (Lipid droplets) |
| Osteogenesis | 3-5 x 10³ cells/cm² | Dexamethasone, Ascorbic Acid, β-Glycerophosphate | 21-28 days | Alizarin Red S (Calcium deposits) |
| Chondrogenesis | 2.5-5 x 10⁵ cells/pellet (Micromass) | TGF-β, Dexamethasone, Ascorbic Acid, Proline | 21-28 days | Alcian Blue (Sulfated proteoglycans) |
Detailed Methodology
- Adipogenic Differentiation: Seed MSCs in growth medium. After 24 hours, switch to adipogenic induction medium (e.g., DMEM with 10% FBS, 1 µM dexamethasone, 0.5 mM IBMX, 100 µM indomethacin, and 10 µg/mL insulin). Differentiate for 14-21 days. Fix cells with 4% PFA and stain with Oil Red O to visualize intracellular lipid vacuoles [25] [69].
- Osteogenic Differentiation: Seed MSCs and culture in osteogenic induction medium (e.g., DMEM with 10% FBS, 50 µM ascorbic acid-2 phosphate, 10 mM β-glycerophosphate, and 0.1 µM dexamethasone) for 21-28 days. Fix cells and stain with Alizarin Red S to detect extracellular matrix calcification [25].
- Chondrogenic Differentiation: Pellet 2.5x10^5 MSCs by centrifugation. Culture the pellet in chondrogenic induction medium (e.g., DMEM with TGF-β, 100 nM dexamethasone, 50 µg/mL ascorbic acid-2 phosphate, and 40 µg/mL proline) for 21-28 days. Fix the pellet, embed in paraffin, section, and stain with Alcian Blue to visualize sulfated glycosaminoglycans in the cartilage matrix [69].
Quantification of Differentiation
While qualitative assessment of staining is common, quantitative methods significantly enhance objectivity and robustness. Color deconvolution followed by digital image analysis provides a straightforward and reproducible quantification method [69].
- Image Acquisition: Capture high-resolution brightfield images of stained cultures using a standardized microscope setup.
- Color Deconvolution: Use the Color Deconvolution plug-in in ImageJ/Fiji to separate the specific stain (e.g., red for Oil Red O, blue for Alcian Blue) from the counterstain (e.g., hematoxylin).
- Threshold and Area Calculation: Apply a consistent threshold to the deconvoluted stain-specific image to create a binary mask. Measure the percentage area covered by the positive signal.
- Calculate Differentiation Ratio (DR): To normalize for cell number, also measure the area of the nuclear counterstain. The Differentiation Ratio (DR) is calculated as: DR = (Area % of differentiation signal) / (Area % of nuclear signal) [69]. This ratio allows for a more accurate comparison of differentiation efficiency across different samples and donors.
The Scientist's Toolkit: Essential Research Reagents
The following table lists key reagents and their functions for the successful execution of MSC characterization protocols.
Table 3: Key Reagent Solutions for MSC Characterization
| Reagent / Kit | Function / Application | Specific Example / Component |
|---|---|---|
| Collagenase Type I/IA | Enzymatic digestion for primary MSC isolation from tissues like adipose tissue and bone marrow [25] [8]. | Preparation of Stromal Vascular Fraction (SVF) from adipose tissue [25]. |
| Liberase | High-purity enzyme blend for efficient tissue dissociation and high MSC yield [8]. | Isolation of bovine adipose tissue-derived MSCs [8]. |
| Fluorochrome-conjugated Antibodies | Immunophenotyping of MSCs via flow cytometry. | Antibodies against CD73, CD90, CD105 (positive) and CD34, CD45 (negative) [3] [69]. |
| Trilineage Differentiation Kits | Standardized media formulations for induced differentiation into adipogenic, osteogenic, and chondrogenic lineages. | Commercial kits containing pre-mixed induction and maintenance media [70]. |
| Oil Red O Solution | Histological staining for detecting intracellular lipid accumulation in adipocytes. | Staining after 21-day adipogenic induction [25] [69]. |
| Alizarin Red S Solution | Histological staining for detecting calcium deposits in osteocytes. | Staining after 21-28 day osteogenic induction [25] [69]. |
| Alcian Blue Solution | Histological staining for detecting sulfated proteoglycans in chondrocytes. | Staining of chondrogenic micromass pellet sections [69]. |
Flow cytometry and trilineage differentiation are non-negotiable, complementary techniques for the definitive characterization of MSCs following isolation. The integration of quantitative methods, such as robust gating strategies in flow cytometry and digital image analysis for differentiation assays, moves beyond qualitative confirmation. It provides a rigorous, data-driven foundation for validating the success of enzymatic isolation protocols, ensuring that the MSC populations used in downstream research and development are well-defined and functionally competent. This rigorous approach is essential for advancing reproducible and clinically relevant applications in regenerative medicine.
The isolation of Mesenchymal Stromal Cells (MSCs) is a critical first step in realizing their potential for regenerative medicine and therapeutic applications. The choice of isolation technique significantly influences the yield, purity, and biological characteristics of the resulting cells, thereby impacting downstream experimental and clinical outcomes. The two predominant methods for isolating MSCs from tissues such as Wharton's jelly (WJ) are enzymatic digestion and the explant culture method. This application note provides a direct comparison of these techniques, framing the analysis within the broader context of mesenchymal stromal cell isolation enzymatic digestion protocol research. Designed for researchers, scientists, and drug development professionals, this document synthesizes current data and provides detailed protocols to inform laboratory decision-making.
Quantitative Comparison: Enzymatic Digestion vs. Explant Method
The following tables summarize key quantitative differences between the two isolation methods, drawing from comparative studies.
Table 1: Comparison of Cell Yield and Growth Kinetics
| Parameter | Enzymatic Digestion Method | Explant Culture Method | References |
|---|---|---|---|
| Primary Cell Yield (P0) | 1.75 ± 2.2 × 10⁵ cells/g | 4.89 ± 3.2 × 10⁵ cells/g (P=0.01) | [7] |
| Time to Confluence | ~7 days | Up to 15 days | [48] |
| Population Doubling Time | Not specified | Shorter doubling times | [48] |
| Initial Cell Population | Heterogeneous, takes longer to enrich for MSCs | More homogenous, purer MSC populations | [65] [71] |
Table 2: Comparison of Cell Characteristics and Practical Considerations
| Aspect | Enzymatic Digestion Method | Explant Culture Method | References |
|---|---|---|---|
| bFGF Release | Lower levels in supernatant | Higher levels released in the first week (55.0 ± 25.6 ng/g total) | [7] |
| Gene Expression | Standard expression profiles | Upregulation of genes related to mitosis and pluripotency markers (OCT4, SOX2, NANOG) | [7] [71] |
| Cell Damage Risk | Higher risk due to potential over-digestion | Lower risk, no enzymatic exposure | [48] |
| Cost & Complexity | Higher (cost of enzymes, growth factors) | Lower (no enzymes or supplementary factors needed) | [48] |
| Ideal Application | Projects requiring rapid cell expansion | Creating cell banks, maintaining genetic stability | [48] |
Detailed Experimental Protocols
Protocol 1: Explant Culture Method for Wharton's Jelly MSC Isolation
This protocol is adapted from methodologies proven to generate homogenous MSC populations with high yield [71] [34].
Sample Preparation:
- Obtain informed consent and ethical approval. Transport the umbilical cord in sterile PBS supplemented with antibiotics (e.g., 1% Penicillin/Streptomycin/Amphotericin B) and process within 3-4 hours of delivery [71] [34].
- Wash the cord thoroughly in PBS to remove residual blood. Cut into 2-3 cm segments.
- Dissect the segments longitudinally and carefully remove the two arteries and one vein using forceps and a scalpel.
- Separate the Wharton's jelly (the loose, mucoid connective tissue) from the denser subamnion membrane. Cut the Wharton's jelly into small explants of approximately 2-3 mm² to 10 mm² [7] [71].
Primary Explant Culture:
- Place the explants directly onto the surface of a tissue culture dish (e.g., 6-well plate). Allow them to adhere to the plastic for 5-10 minutes at room temperature without allowing them to dry out [71].
- Gently add a low volume of complete culture medium, such as Dulbecco's Modified Eagle's Medium (DMEM, low glucose) supplemented with 10-15% Fetal Bovine Serum (FBS) and 1% L-glutamine and antibiotics. Ensure the medium level does not submerge the explants completely to prevent detachment [71] [34].
- Incubate the culture at 37°C in a humidified atmosphere of 5% CO₂. Do not move the dish for the first 5-7 days to allow for stable cell migration.
- After 2 days, perform a partial medium change. Thereafter, change the medium twice weekly.
- Fibroblast-like, adherent cells will begin to migrate from the tissue explants within 3-7 days. The explants can be carefully removed after 10-14 days, and the adherent cells can be cultured further until they reach 70-80% confluence, which is designated as passage 0 (P0) [7] [34].
Protocol 2: Enzymatic Digestion Method for Wharton's Jelly MSC Isolation
This protocol utilizes proteolytic enzymes to dissociate tissue and is valued for its rapidity [7] [3].
Tissue Digestion:
- After washing and dissecting the umbilical cord as described in Protocol 1, mine the Wharton's jelly into very small pieces.
- Transfer the minced tissue to a digestion solution. A common and effective enzyme is 0.1% Collagenase type II in serum-free media, though other options like Liberase can be optimized for yield [7] [8].
- Incubate the tissue-enzyme mixture for 30 minutes to 3 hours at 37°C with gentle agitation. The incubation time and enzyme concentration should be optimized to balance yield and viability, avoiding over-digestion [7] [8].
- Neutralize the enzymatic reaction by adding complete culture medium containing FBS.
Cell Seeding and Culture:
- Filter the resulting cell suspension through a cell strainer (e.g., 70-100 µm) to remove undigested tissue fragments.
- Centrifuge the filtrate to pellet the cells. Wash the pellet with PBS and re-centrifuge.
- Resuspend the final cell pellet in complete culture medium (e.g., DMEM with 10% FBS) and seed them into an appropriate tissue culture flask.
- Incubate the culture at 37°C and 5% CO₂, changing the medium twice weekly.
- The primary culture (P0) is typically ready for passaging in about 7 days, though initial populations may be heterogeneous and require further passages to enrich for MSCs [48] [65].
Workflow and Signaling Pathway Diagrams
Diagram 1: MSC Isolation Workflow Comparison
This diagram illustrates the procedural differences and outcomes between the two primary isolation methods.
Diagram 2: bFGF Signaling and Mitogenic Pathway in Explant-Derived MSCs
This diagram outlines the signaling pathway by which explant-derived MSCs may exhibit enhanced proliferative characteristics, linked to the observed upregulation of mitotic genes and release of basic Fibroblast Growth Factor (bFGF) [7].
The Scientist's Toolkit: Key Research Reagent Solutions
The following table details essential materials and reagents used in the featured isolation protocols, with explanations of their critical functions.
Table 3: Essential Reagents for MSC Isolation from Wharton's Jelly
| Reagent / Material | Function in Protocol | Examples / Notes |
|---|---|---|
| Collagenase Type I/II/IV | Proteolytic enzyme for digesting collagen in the extracellular matrix during enzymatic isolation. | Concentration and type (e.g., Liberase vs. Collagenase) must be optimized for specific tissue and yield goals [7] [8]. |
| Dulbecco's Modified Eagle's Medium (DMEM) | Basal nutrient medium providing essential vitamins, amino acids, and energy for cell growth. | Often used in low-glucose formulation. Can be supplemented with DMEM-F12 for optimized growth [7] [34]. |
| Fetal Bovine Serum (FBS) | Provides a complex mixture of growth factors, hormones, and attachment factors crucial for cell adhesion and proliferation. | Typically used at 10-15% concentration. Serum quality and batch are critical for consistency [71] [34]. |
| Basic Fibroblast Growth Factor (bFGF) | A supplement that significantly enhances MSC proliferation rates and helps maintain stemness. | Used at 10 ng/ml. Particularly beneficial for expanding cultures after initial isolation [34]. |
| Trypsin-EDTA | A proteolytic enzyme used for passaging adherent cells by detaching them from the culture surface. | Standard concentration is 0.25%. Exposure time should be minimized to maintain cell viability [71] [34]. |
| CellBIND Surface or Equivalent | A specially treated tissue culture plastic surface that enhances cell attachment and growth. | Particularly valuable for improving initial cell attachment in both enzymatic and explant methods, boosting overall yield [48]. |
The choice between enzymatic digestion and explant culture for MSC isolation is not a matter of one being universally superior, but rather depends on the specific goals and constraints of the research or therapeutic project. The explant method is advantageous for generating a purer, more homogeneous population of MSCs with a higher initial yield, superior genetic stability, and the benefit of an active extracellular matrix that releases growth factors like bFGF. In contrast, the enzymatic method provides speed, with cells reaching confluence more rapidly, which can be critical in time-sensitive therapeutic applications, though it carries a risk of cell damage and requires more expensive reagents. Researchers must weigh these factors—yield, purity, growth kinetics, cost, and intended application—to select the most appropriate isolation technique for their work in mesenchymal stromal cell research.
Mesenchymal stromal cells (MSCs) represent a cornerstone of regenerative medicine, with their therapeutic potential extensively explored for a diverse range of human diseases [72]. While MSCs share fundamental biological properties, their functional characteristics, differentiation potential, and secretory profiles exhibit significant variation depending on their tissue of origin [28] [43]. Understanding these source-dependent variations is critical for selecting the optimal MSC type for specific therapeutic applications and for developing standardized isolation protocols that account for intrinsic biological differences. This Application Note provides a detailed comparative analysis of MSCs derived from three clinically relevant sources: adipose tissue (AD-MSCs), dental pulp (DP-MSCs), and umbilical cord (UC-MSCs), focusing on their molecular signatures, functional capabilities, and isolation methodologies within the context of enzymatic digestion protocol research.
Molecular and Functional Characterization
Transcriptomic and Phenotypic Heterogeneity
Comprehensive transcriptome analyses reveal fundamental differences in gene expression patterns among MSCs derived from various tissues. A 2023 study demonstrated that while UC-MSCs show the strongest correlation with bone marrow MSCs (BM-MSCs), all MSC sources exhibit distinct transcriptional profiles [73].
Key findings include:
- Lower differentially expressed genes in BM-MSCs, DP-MSCs, and AD-MSCs compared to UC-MSCs were predominantly enriched in actin-related processes
- Higher differentially expressed genes were primarily involved in immunological functions
- CD200 (FPKM >10) was exclusively detected in UC-MSCs
- CD106 was present in AD-MSCs and DP-MSCs (FPKM >10) but not in UC-MSCs [73]
These findings suggest that CD200 and CD106 could serve as benchmark molecules to monitor MSC proliferation and differentiation potential, providing valuable quality control markers for therapeutic applications.
Table 1: Molecular Characteristics of MSCs from Different Sources
| Characteristic | AD-MSCs | DP-MSCs | UC-MSCs |
|---|---|---|---|
| Unique Marker Expression | CD106+ | CD106+ | CD200+ |
| Transcriptomic Profile | Enriched in actin-related terms | Enriched in actin-related terms | Strong correlation with BM-MSCs |
| Immunological Processes | Higher differentially expressed genes | Higher differentially expressed genes | Distinct immunologic enrichment |
| Size & Morphology | Larger cells | Consistently smaller, Nestin-positive [43] | Not specified |
Differentiation Potential and Functional Heterogeneity
The trilineage differentiation capacity—osteogenic, chondrogenic, and adipogenic—varies considerably among MSC sources, influencing their suitability for specific regenerative applications.
Notable differences include:
- DP-MSCs demonstrate superior osteogenic capacity but a weaker adipogenic potential compared to BM-MSCs [74]
- DP-MSCs consistently show higher proliferation rates than AD-MSCs [43]
- DP-MSCs were unable to perform adipogenesis in trilineage differentiation assays, despite possessing other typical MSC characteristics [43]
- AD-MSCs exhibit robust adipogenic differentiation capability, consistent with their tissue origin [75]
These functional specializations align with the developmental origins and physiological roles of the source tissues, highlighting the importance of matching MSC source to therapeutic intent.
Secretome Composition and Extracellular Vesicle Profiles
The secretome—comprising soluble factors and extracellular vesicles (EVs)—is increasingly recognized as the primary mediator of MSC therapeutic effects, and its composition varies significantly by source.
Comparative analyses reveal:
- AD-MSCs and DP-MSCs release a comparable number of EVs, but AD-MSCs produce significantly more smaller exosomes [43]
- MSC populations release tissue-specific sets of microRNAs, either free or enclosed in EVs:
- DP-MSC-derived microRNAs are primarily involved in oxidative stress and apoptosis pathways
- AD-MSC-derived microRNAs play regulatory roles in cell cycle and proliferation [43]
- Secretome analysis shows significant variations in anti-inflammatory and pro-inflammatory cytokines, chemokines, and growth factors between MSC sources [43]
These source-dependent secretome variations have profound implications for cell-free therapeutic applications, suggesting that specific MSC types may be preferable for particular disease targets.
Therapeutic Efficacy in Disease Models
Performance in Osteoporosis Treatment
A 2024 study directly compared the therapeutic efficacy of MSCs from different sources in treating postmenopausal osteoporosis using an ovariectomy (OVX)-induced mouse model [74] [76]. The findings demonstrated substantial differences in bone regeneration capacity.
Table 2: Therapeutic Efficacy of Different MSCs in Osteoporosis Model
| Parameter | AD-MSCs | DP-MSCs | AM-MSCs | UC-MSCs |
|---|---|---|---|---|
| Trabecular Bone Volume | Moderate improvement | Highest improvement | Moderate improvement | Moderate improvement |
| Bone Mineral Density | Moderate improvement | Highest improvement | Moderate improvement | Moderate improvement |
| Immunoregulatory Capacity | Moderate | Superior (regulated Th17/Treg and M1/M2 ratios) | Moderate | Moderate |
| Osteoblastogenesis | Moderate | Enhanced | Moderate | Moderate |
| Bone Resorption Inhibition | Moderate | Most effective | Moderate | Moderate |
Key mechanistic insights:
- DP-MSC infusion maintained trabecular bone mass more efficiently with corresponding improvements in trabecular bone parameters [74]
- DP-MSCs exhibited greater immunoregulatory capabilities, effectively regulating the Th17/Treg and M1/M2 ratios [74]
- The superior performance of DP-MSCs highlights how source-specific properties significantly influence therapeutic outcomes in bone regeneration applications
Isolation and Expansion Methodologies
Robust and standardized isolation methods are essential for minimizing technical variability and ensuring reproducible MSC characteristics across different donors and studies.
Adipose-Derived MSCs (AD-MSCs)
AD-MSCs can be isolated through two primary approaches, each with distinct advantages:
Enzymatic Digestion (Stromal Vascular Fraction - SVF):
- Abdominal adipose tissue is washed with DPBS and subjected to overnight enzymatic digestion at 37°C with collagenase 1A [43]
- The digested material is centrifuged (10' at 1200 g), and the cell pellet is plated onto tissue culture dishes [43]
- This method yields cells termed ADSCs-SVF
Mechanical Fragmentation (MF):
- The LIPOGEMS system separates lipoaspirate into three layers: oil (top), intact adipose tissue (middle), and cellular contaminants (bottom) [43]
- The middle layer is placed in αMEM supplemented with 20% FBS [43]
- ADSCs (termed ADSCs-MF) outgrow from fragments floating on the medium surface over 2 weeks [43]
A 2023 study introduced a novel enzyme-free mechanical approach using a 'microlyzer' device, which fragments adipose tissue into microparticles through blades of varying diameters (2400, 1200, 600 μm) [77]. This system produces SVF with progenitor cell counts comparable to enzymatic methods while avoiding enzyme-related safety concerns [77].
Dental Pulp-Derived MSCs (DP-MSCs)
DP-MSCs are typically isolated via mechanical fragmentation from open apex third molars [43]:
- Teeth are cut at the amelo-cement junction with a diamond disc, and pulp is gently removed using a sterile dental scalpel [43]
- Each pulp is divided into coronal and radicular portions, fragmented into small pieces (1-2 mm³) [43]
- Fragments are washed by centrifugation (300 g for 5'), seeded onto tissue culture plates [43]
- Cells outgrow from fragments after 2-4 weeks, forming a monolayer that reaches 80% confluence [43]
- Regional specialization is observed between coronal pulp stromal cells (CPSCs) and radicular pulp stromal cells (RPSCs) [43]
Umbilical Cord-Derived MSCs (UC-MSCs)
A robust standardized method for hUC-MSC isolation, validated across 90 donors, involves several critical steps [27]:
- UC collection can occur up to 6 h after birth, with processing initiation possible up to 48 h after collection without impacting hUC-MSC characteristics [27]
- Removal of blood vessels before explant cultures improves hUC-MSC purity [27]
- Expansion in Minimum Essential Medium α supplemented with human platelet lysate increases reproducibility compared to Dulbecco's Modified Eagle's Medium with fetal bovine serum [27]
- Isolated hUC-MSCs show ~98.9% purity, >97% viability, and high proliferative capacity [27]
Research Reagent Solutions
Table 3: Essential Materials for MSC Isolation and Characterization
| Reagent/Consumable | Function/Application | Examples/Specifications |
|---|---|---|
| Collagenase 1A | Enzymatic digestion of adipose tissue for SVF isolation | Sigma-Aldrich; overnight digestion at 37°C [43] |
| αMEM Medium | Basal medium for MSC expansion | Aurogene; supplemented with FBS and antibiotics [43] |
| Fetal Bovine Serum (FBS) | Serum supplement for cell culture | Gibco; 10-20% concentration [43] |
| Trypsin-EDTA | Cell detachment from culture vessels | Sigma-Aldrich; 5 min incubation at 37°C [43] |
| Human Platelet Lysate | Serum alternative for UC-MSC expansion | Superior reproducibility for UC-MSC expansion [27] |
| Microlyzer Device | Mechanical fragmentation of adipose tissue | ISO 13485 certified, CE marked; blades of 2400, 1200, 600 μm [77] |
| Diamond Disc | Tooth sectioning for DP-MSC isolation | For cutting at amelo-cement junction [43] |
| Percoll | Density gradient centrifugation | MSC isolation alternative [28] |
Experimental Workflows and Signaling Pathways
MSC Isolation and Characterization Workflow
The following diagram illustrates the core experimental workflow for isolating and characterizing MSCs from different tissue sources, integrating key decision points and characterization steps:
MSC Signaling and Therapeutic Action Pathways
The therapeutic effects of MSCs are mediated through complex signaling pathways and secretory profiles that vary by source. The following diagram illustrates key mechanistic pathways:
The source-dependent variations among AD-MSCs, DP-MSCs, and UC-MSCs extend beyond mere tissue origin to encompass fundamental differences in transcriptomic profiles, differentiation potentials, secretome compositions, and therapeutic efficacies. DP-MSCs demonstrate particular promise for bone-related regeneration, while UC-MSCs offer advantages in proliferation and immunomodulation. AD-MSCs remain valuable for their accessibility and adipogenic potential. The selection of an appropriate MSC source must be guided by the specific therapeutic application, with careful consideration of the isolation and expansion methods that best preserve the desired functional characteristics. Standardized protocols that account for these source-specific variations will enhance reproducibility and clinical translation of MSC-based therapies.
Within the broader scope of a thesis investigating enzymatic digestion protocols for the isolation of Mesenchymal Stromal Cells (MSCs), this document details subsequent critical application notes for characterizing the functional potency of the obtained cells. The therapeutic potential of MSCs, whether used directly or in cell-free therapies, hinges primarily on their immunomodulatory capacity, which is largely mediated by their secretome—a complex mixture of soluble factors and extracellular vesicles (EVs) [78]. Therefore, standardizing robust functional potency assays is indispensable for correlating specific isolation methods with therapeutic efficacy and ensuring batch-to-batch consistency in research and drug development.
This protocol provides a detailed framework for assessing the immunomodulatory capacity of MSCs through secretome analysis, focusing on two key functional readouts: the suppression of innate immune responses and the inhibition of T-cell proliferation.
Experimental Workflow & Signaling Pathways
The following sections provide detailed protocols for the key experiments in this workflow.
Key Signaling Pathways in MSC Licensing
The immunomodulatory phenotype of MSCs is not constitutive but is induced through a process called "licensing" using pro-inflammatory cytokines. The optimized protocol below uses a combination of IFN-γ and TNF-α, which activate synergistic intracellular signaling pathways to enhance the production of immunosuppressive molecules [79].
A comprehensive potency assessment involves a sequence of steps from MSC culture and licensing to secretome collection, fractionation, and functional analysis. The workflow below integrates these key stages systematically.
Detailed Experimental Protocols
Protocol 1: MSC Licensing and Secretome Production
This protocol outlines the optimized procedure for licensing MSCs to enhance their immunomodulatory phenotype and for collecting the resulting secretome [79].
- Cell Culture: Maintain immortalized human adipose tissue-derived MSCs (e.g., hTERT-AT-MSCs, ASC52telo) in complete MSC basal medium. Culture cells at 37°C in a humidified atmosphere of 5% CO₂. Use cells between passages 4 and 7 for experiments to ensure consistency [79].
- Licensing Cocktail Preparation: Prepare a 1:1 ratio of IFN-γ and TNF-α in fresh basal medium. The optimal final concentration for each cytokine is 60 ng/mL (i.e., 60 ng/mL IFN-γ + 60 ng/mL TNF-α) [79].
- Licensing Induction: When cultures reach 90% confluence, replace the growth medium with the licensing cocktail. Incubate the cells for 24 hours (overnight) [79].
- Secretome Collection (Conditioned Media): After the licensing period, carefully wash the cells with PBS to remove all cytokines. Add fresh, serum-free basal medium and incubate for 48 hours for secretome production [79].
- Clarification: Collect the conditioned media and centrifuge at 300 × g for 10 minutes to remove any detached cells. Then, clarify the supernatant by passing it through a 0.2 µm filter. This yields the "Clarified Secretome" [78]. Aliquot and store at -80°C.
Protocol 2: Secretome Fractionation
The clarified secretome can be processed further to isolate specific fractions, allowing for a more detailed analysis of the active components [78].
- Tangential Flow Filtration (TFF): To concentrate the secretome and separate components by size, use a TFF system with membranes of varying molecular weight cutoffs (e.g., 5, 10, 30, or 100 kDa). Diafilter the clarified secretome against PBS. The resulting "Concentrated Secretome" is enriched with components larger than the chosen cutoff [78].
- Ultracentrifugation: To separate soluble factors from extracellular vesicles (EVs), subject the clarified secretome to ultracentrifugation at 150,000 × g for 2 hours at 4°C [78].
- Carefully collect the supernatant, which constitutes the "Soluble Factors Fraction."
- Resuspend the pellet (containing the EV fraction) in an appropriate volume of PBS.
- Storage: All fractionated samples (TFF concentrates, soluble factors, and EVs) should be aliquoted and stored at -80°C until further use.
Protocol 3: Functional Potency Assays
Innate Immunomodulation Assay (NF-κB/IRF Pathway)
This assay evaluates the secretome's ability to suppress activation of innate immune pathways [78].
- PBMC Activation: Isolate human Peripheral Blood Mononuclear Cells (PBMCs) from whole blood using density gradient centrifugation. Activate the PBMCs by treating them with resiquimod (a TLR7/8 agonist) for the duration of the co-culture.
- Co-culture with Secretome: Co-culture the activated PBMCs with the various secretome fractions (clarified, concentrated, or soluble factors). Use a range of dilutions (e.g., 1:2 to 1:100) to assess dose-dependent effects.
- Reporter Assay: After co-culture, collect the PBMC supernatant and transfer it to THP-1 Dual reporter cells. This cell line is engineered to produce a measurable signal (e.g., secreted embryonic alkaline phosphatase, SEAP) upon activation of the NF-κB and IRF pathways.
- Measurement: Quantify the reporter signal according to the manufacturer's instructions. A reduction in signal compared to the activated control indicates that the secretome fraction has inhibited the NF-κB and/or IRF pathways.
T-cell Proliferation Assay
This assay measures the secretome's capacity to suppress adaptive immune responses by inhibiting T-cell division [78].
- T-cell Labeling: Isolate PBMCs as above. Label the cells with a fluorescent cell division tracker dye, such as CFSE (Carboxyfluorescein succinimidyl ester) or an equivalent dye. The dye dilutes by approximately half with each cell division.
- T-cell Activation: Activate the labeled PBMCs using a mitogen such as Phytohemagglutinin (PHA) in combination with IL-2 (e.g., 5 µg/mL PHA + 20 IU/mL IL-2) [78].
- Co-culture with Secretome: Culture the activated, labeled PBMCs in the presence of the different secretome fractions.
- Flow Cytometry Analysis: After 3-5 days, harvest the cells and analyze them by flow cytometry. Gate on CD3⁺ T-cells and measure the dilution of the fluorescent dye.
- Data Analysis: A reduction in dye dilution (i.e., fewer cell divisions) in treated samples compared to the activated control indicates suppression of T-cell proliferation. Data can be reported as percentage inhibition or proliferation index.
Quantitative Data and Analysis
The immunomodulatory effects of the MSC secretome are multifaceted, with different fractions mediating suppression through distinct pathways. The table below summarizes the key findings from the functional assays [78].
Table 1: Functional Effects of Different MSC Secretome Fractions
| Secretome Fraction | Effect on NF-κB/IRF Pathway (Innate Immunity) | Effect on T-cell Proliferation (Adaptive Immunity) | Key Mediators |
|---|---|---|---|
| Clarified Secretome | Strong, dose-dependent inhibition [78] | Minimal effect [78] | Soluble factors < 5 kDa (e.g., PGE₂) [78] |
| TFF-Concentrated Secretome | Reduced inhibitory effect [78] | Strong, dose-dependent inhibition [78] | Components > 100 kDa [78] |
| Soluble Factors (Ultracentrifugation Supernatant) | Strong, dose-dependent inhibition [78] | Data not available in search results | Soluble factors < 5 kDa (e.g., PGE₂, Kynurenine) [78] |
| EV Pellet (Ultracentrifugation) | Lost inhibitory effect [78] | Data not available in search results | Not the primary mediators of innate pathway suppression [78] |
Optimized Licensing Protocol Parameters
Systematic optimization of the licensing protocol is critical for maximizing the potency of the resulting secretome. The following table outlines the key parameters and their optimized values based on recent research [79].
Table 2: Optimized Parameters for MSC Licensing with IFN-γ and TNF-α
| Parameter | Options Evaluated | Optimized Condition | Impact on Secretome Potency |
|---|---|---|---|
| Cytokine Ratio | 1:1, 2:1, 1:2 (IFN-γ:TNF-α) [79] | 1:1 Ratio [79] | Synergistic activation of JAK/STAT and NF-κB pathways [79] |
| Cytokine Concentration | 0 - 100 ng/mL (total) [79] | 60 ng/mL each (120 ng/mL total) [79] | Maximizes immunomodulatory factor production (e.g., Gal-9, IDO) without inducing toxicity [79] |
| Licensing Duration | 1 - 3 days [79] | 24 hours (Overnight) [79] | Sufficient to induce a robust MSC2 phenotype [79] |
| Production Confluence | 60% - 90% [79] | 90% Confluence [79] | Higher cell density yields an optimized conditioned media with enhanced immunomodulatory properties [79] |
| Secretome Production Time | 24, 48, 72 hours [79] | 48 hours [79] | Balances high yield of bioactive factors with cell viability [79] |
The Scientist's Toolkit: Essential Research Reagents
A successful potency assay relies on a set of well-defined reagents and tools. The following table lists key materials cited in the protocols.
Table 3: Essential Reagents and Materials for Secretome Potency Analysis
| Reagent/Material | Function/Description | Example/Reference |
|---|---|---|
| IFN-γ & TNF-α | Pro-inflammatory cytokines used to license MSCs and enhance their immunomodulatory phenotype. | Recombinant Human proteins [79] |
| hTERT-AT-MSCs | A standardized, immortalized human adipose tissue-derived MSC line; reduces donor variability. | ASC52telo (ATCC SCRC-4000) [79] |
| Tangential Flow Filtration (TFF) | A filtration method for concentrating and diafiltering the secretome based on molecular weight cutoffs. | Systems with 5, 10, 30, 100 kDa membranes [78] |
| Liberase | A highly purified enzyme blend for efficient isolation of MSCs from adipose tissue, maximizing cell yield. | 0.1% for 3 hours incubation [8] |
| THP-1 Dual Cells | A reporter cell line used to quantify activation of the NF-κB and IRF pathways in innate immunomodulation assays. | Invivogen [78] |
| CFSE | A fluorescent dye used to track and quantify cell proliferation in the T-cell suppression assay. | Carboxyfluorescein succinimidyl ester [78] |
| PHA & IL-2 | Mitogen and cytokine combination used to activate T-cells and induce proliferation in functional assays. | e.g., 5 µg/mL PHA + 20 IU/mL IL-2 [78] |
| MACSPlex Kits | Flow cytometry-based kits for the phenotyping and characterization of extracellular vesicles (EVs). | Miltenyi Biotec [78] |
| ELISA Kits | For quantifying specific soluble factors in the secretome, such as PGE₂ and kynurenine. | Cayman Chemical, Immusmol [78] |
Impact of Isolation Method on Final Cell Product for Clinical Applications
The transition of Mesenchymal Stromal Cell (MSC)-based therapies from research to clinical applications demands rigorous standardization of manufacturing processes. The initial isolation method is a critical determinant of the final cell product's characteristics, influencing yield, purity, functional properties, and ultimately, therapeutic efficacy [80]. As MSC-based Advanced Therapy Medicinal Products (ATMPs) progress through clinical trials—with over 1400 studies registered and 11 products currently licensed—the need for robust, reproducible isolation protocols becomes paramount for regulatory approval and clinical success [80]. This application note examines how enzymatic digestion and explant culture methods impact critical quality attributes of MSCs from various tissue sources, providing structured protocols and analytical frameworks to guide therapeutic development.
Comparative Analysis of MSC Isolation Techniques
Technical Principles and Procedural Comparison
The two primary methods for MSC isolation—enzymatic digestion and explant culture—operate on fundamentally different principles with distinct procedural implications:
Enzymatic Digestion: Utilizes proteolytic enzymes (e.g., collagenase, liberase, trypsin) to chemically degrade the extracellular matrix, liberating individual cells into suspension for immediate plating [48]. This method actively disrupts tissue architecture and cell-cell interactions.
Explant Culture: Relies on the innate migratory capacity of MSCs, which physically emigrate from tissue fragments plated on culture surfaces without enzymatic assistance [48]. This approach preserves native cell interactions and matrix components.
Table 1: Fundamental Characteristics of MSC Isolation Techniques
| Parameter | Enzymatic Digestion | Explant Culture |
|---|---|---|
| Principles | Uses proteolytic enzymes (e.g., collagenase, liberase) to digest extracellular matrix and release individual cells [48] | Relies on cell migration from tissue fragments plated directly on culture surfaces without enzymes [48] |
| Procedural Time | Rapid cell release (hours); confluence typically achieved in ~7 days [48] | Slow cell emigration (days to weeks); confluence typically achieved in ~15 days [48] [32] |
| Technical Complexity | Requires optimization of enzyme type, concentration, and incubation time; neutralization steps needed [8] | Technically straightforward with minimal handling steps; reduced optimization parameters [27] |
| Automation Potential | Highly amenable to closed-system automation and standardization [32] | Limited automation potential due to manual tissue handling and fragmentation [27] |
| Regulatory Considerations | Enzymes require qualification/validation as critical reagents; potential animal-origin concerns [80] | Reduced reagent complexity; simplified regulatory documentation [27] |
Impact on Critical Quality Attributes of MSC Products
Isolation methodology significantly influences multiple critical quality attributes (CQAs) of the resulting MSC products, with implications for their therapeutic profile:
Table 2: Impact of Isolation Method on MSC Product Characteristics
| Cell Product Attribute | Enzymatic Digestion | Explant Culture | Clinical Significance |
|---|---|---|---|
| Initial Cell Yield | Variable by tissue and protocol: 30.48-67.1 × 106 cells/g AT (optimal conditions) [8]; 1.75 ± 2.2 × 105/g WJ [7] | Generally lower initial yield: 4.89 ± 3.2 × 105/g WJ [7] | Determines starting biomass and expansion requirements; impacts cost-of-goods |
| Cell Viability | Potentially compromised by over-digestion (>95% with optimized protocols) [8] | Typically high viability with minimal processing stress [48] | Affects expansion potential and product consistency |
| Population Doubling Time | Varies with protocol: ~2.76 days for UC-MSC [32] | Generally shorter doubling times [48] | Impacts production timeline and cellular age at harvest |
| Cellular Senescence | Potential for replicative stress due to enzymatic exposure | Reduced processing-induced stress [48] | Affects in vivo persistence and therapeutic durability |
| Surface Marker Expression | Potential alteration of epitopes by enzyme activity [48] | Native marker presentation preserved [48] | Impacts homing, potency, and product characterization |
| Secretory Profile | Altered paracrine signature due to enzymatic stress | Enhanced growth factor release (e.g., bFGF: 55.0 ± 25.6 ng/g in explant culture) [7] | Critical for paracrine-mediated therapeutic mechanisms |
| Genetic Stability | Potential for enzyme-induced stress responses | Maintained genetic stability with minimal biological drift [48] | Safety consideration for clinical applications |
| Differentiation Potential | Protocol-dependent: maintained in optimized isolations [8] [32] | Preserved differentiation capacity [25] | Important for tissue-specific applications |
The explant method demonstrates particular advantages for secretory function, with studies showing it releases significantly higher levels of basic Fibroblast Growth Factor (bFGF) in supernatant media during the first week of culture compared to enzymatic digestion (total bFGF release of 55.0 ± 25.6 ng/g tissue) [7]. Additionally, explant-derived MSCs show upregulation of genes related to mitosis, suggesting enhanced proliferative capacity in the initial phases of culture [7].
Tissue-Specific Optimization Requirements
Source-Dependent Methodological Considerations
Different tissue sources present unique challenges and requirements for MSC isolation, necessitating tailored approaches:
Adipose Tissue: Requires efficient disruption of dense lipid-rich matrix. Liberase TM at 0.1% for 3 hours demonstrated superior yield (30.48-67.1 × 106 cells/g tissue) while maintaining differentiation potential and surface marker expression [8]. Collagenase type I remains frequently used but with variable efficiency across donors [8].
Umbilical Cord/Wharton's Jelly: Rich in mucopolysaccharides requiring specific enzymatic blends. Explant methods provide excellent yield (4.89 ± 3.2 × 105/g versus 1.75 ± 2.2 × 105/g for enzymatic) with enhanced growth factor retention [7]. Vessel removal before explant culture improves MSC purity [27].
Dental Pulp: Limited tissue volume favors explant methods. Regional compartmentalization (coronal vs. radicular) affects MSC characteristics regardless of isolation method [25].
Comprehensive Experimental Protocols
Optimized Enzymatic Protocol for Adipose Tissue-Derived MSCs
Objective: Isolation of MSCs from bovine subcutaneous adipose tissue with maximum cell yield and maintained functionality for cultured meat production applications [8].
Reagents:
- Liberase TM (0.1% solution in PBS)
- Dulbecco's Modified Eagle Medium (DMEM)
- Fetal Bovine Serum (FBS)
- Antibiotic-Antimycotic solution
- Phosphate Buffered Saline (PBS)
Procedure:
- Tissue Processing: Mince 1g adipose tissue into <2mm fragments using sterile surgical blades.
- Enzymatic Digestion: Incubate tissue fragments with 10mL 0.1% Liberase TM solution at 37°C for 3 hours with gentle agitation.
- Reaction Neutralization: Add equal volume DMEM supplemented with 10% FBS.
- Cell Collection: Filter through 100μm strainer, centrifuge at 300×g for 10 minutes.
- Red Blood Cell Removal: Resuspend pellet in RBC lysis buffer if necessary.
- Plating and Culture: Plate cells at 1×106 cells/cm2 in growth medium.
- Medium Exchange: Replace after 24 hours to remove non-adherent cells, then every 3-4 days.
Validation Parameters:
- Cell yield: >35×106 cells/g tissue at P1
- Viability: >95% by trypan blue exclusion
- Differentiation: Tri-lineage potential confirmed
- Surface markers: CD73+, CD90+, CD105+, CD34-, CD45-
Standardized Explant Protocol for Umbilical Cord-Derived MSCs
Objective: Isolation of high-purity MSCs from human umbilical cord with minimal technical variability for clinical applications [27].
Reagents:
- Minimum Essential Medium α (MEMα)
- Human Platelet Lysate (5%)
- Antibiotic-Antimycotic solution
- Phosphate Buffered Saline (PBS)
Procedure:
- Collection: Process UC within 6 hours of birth, initiate processing within 48 hours.
- Vessel Removal: Dissect and remove umbilical arteries and vein.
- Tissue Preparation: Cut Wharton's jelly into 2-3mm segments.
- Explant Plating: Place segments directly on culture surface without enzymatic treatment.
- Initial Culture: Maintain explants in MEMα with 5% human platelet lysate for 2 weeks.
- Explant Removal: Carefully remove tissue segments after 14 days.
- Cell Expansion: Culture emigrated cells to confluence (additional 7 days).
Critical Control Points:
- UC collection-to-processing interval: ≤6 hours
- Processing-to-culture initiation: ≤48 hours
- FBS-free medium with human platelet lysate enhances reproducibility
- Vessel removal essential for MSC purity (~98.9%)
Validation Parameters:
- Purity: >95% MSC markers, <2% hematopoietic contaminants
- Viability: >97%
- Population doublings: >15 without senescence
- Functional: T-cell proliferation inhibition >50%
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Reagent Solutions for MSC Isolation and Characterization
| Reagent Category | Specific Examples | Function & Application Notes |
|---|---|---|
| Digestive Enzymes | Collagenase Type I [8], Liberase TM [8], Collagenase Type II [7], Trypsin [8] | Tissue-specific efficiency; Liberase TM shows superior yield for adipose tissue [8] |
| Culture Media | Low-Glucose DMEM [7], MEMα [27], αMEM [25] | MEMα with human platelet lysate enhances reproducibility for UC-MSCs [27] |
| Serum Supplements | Fetal Bovine Serum [8], Human Platelet Lysate [27] | Xeno-free alternatives critical for clinical applications |
| Characterization Antibodies | CD73, CD90, CD105 [32], CD34, CD45 [32], HLA-DR [7] | Essential for ISCT phenotype confirmation [32] |
| Differentiation Kits | Osteogenic: Ascorbic acid, β-glycerophosphate, dexamethasone [25] Adipogenic: IBMX, indomethacin, insulin [25] Chondrogenic: TGF-β, BMPs [25] | Standardized kits improve inter-study comparability |
| Cell Attachment Surfaces | Corning CellBIND [48], Standard tissue culture plastic | Enhanced attachment surfaces improve yield for both methods [48] |
Strategic Framework for Isolation Method Selection
Diagram 1: Strategic decision framework for MSC isolation method selection illustrating key considerations including clinical needs, tissue characteristics, and technical constraints that inform method choice between explant culture, enzymatic digestion, or hybrid approaches.
The selection between enzymatic and explant isolation methods represents a foundational decision in MSC product development that extends throughout the therapeutic product's lifecycle. While enzymatic digestion offers advantages in standardization and scalability for large-scale production, explant methods demonstrate benefits in cellular function and phenotype preservation. The optimal approach must be determined through systematic evaluation of target indication, tissue source, and manufacturing constraints, with method-specific protocols rigorously optimized and validated against critical quality attributes. As regulatory scrutiny of ATMPs intensifies, comprehensive documentation of isolation methodology and its impact on product characteristics becomes essential for clinical translation and commercialization success.
Conclusion
The enzymatic digestion protocol is a cornerstone of efficient and reproducible MSC isolation, yet its success hinges on a nuanced, tissue-specific approach. This synthesis underscores that there is no universal method; optimal outcomes require careful selection of enzymes like Liberase™ for adipose tissue and tailored protocols for umbilical cord or dental pulp. Adherence to ISCT characterization criteria and GMP regulations is non-negotiable for clinical translation. While enzymatic digestion often provides superior initial yield and scalability, the explant method can offer advantages in initial cell purity. Future directions must focus on standardizing these protocols, further elucidating how isolation affects long-term MSC function and secretome, and developing serum-free, xeno-free enzymatic cocktails to ensure the safety and efficacy of MSC-based therapies in regenerative medicine.
References
- 1. Mesenchymal Stem Cells: Revisiting History, Concepts, ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC2613570/]
- 2. Mesenchymal stem/stromal cells (MSCs): origin, immune ... [https://www.nature.com/articles/s41423-023-01034-9]
- 3. Validated methods for isolation and qualification of ... [https://translational-medicine.biomedcentral.com/articles/10.1186/s12967-025-06972-8]
- 4. From “stem” to “stromal”: Reframing the nomenclature of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12553919/]
- 5. Mesenchymal stem cell [https://en.wikipedia.org/wiki/Mesenchymal_stem_cell]
- 6. Comparing mechanical and enzymatic isolation procedures to ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11584359/]
- 7. Comparison of Explant-Derived and Enzymatic Digestion ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3638666/]
- 8. Evaluation of enzymatic protocols to optimize efficiency ... [https://www.nature.com/articles/s41538-024-00313-7]
- 9. Defining minimal criteria for peer-reviewed reporting of ... [https://www.sciencedirect.com/science/article/pii/S1465324925007443]
- 10. Validated methods for isolation and qualification of ... [https://pubmed.ncbi.nlm.nih.gov/40898279/]
- 11. Derived from Mesenchymal and Adipose... Stromal Cells Bone Marrow [https://bio-protocol.org/en/bpdetail?id=3534&type=0]
- 12. Isolation of Adipose-Derived Mesenchymal Stromal/Stem ... [https://pubmed.ncbi.nlm.nih.gov/40445288/]
- 13. for Protocol of human isolation and... bone marrow stromal cells [https://pubmed.ncbi.nlm.nih.gov/39813119/]
- 14. A simple and reliable protocol for long-term culture of murine bone ... [https://biologicalproceduresonline.biomedcentral.com/articles/10.1186/s12575-019-0091-3]
- 15. Isolation and Establishment of Mesenchymal Stem Cells ... [https://en.bio-protocol.org/en/bpdetail?id=2735&type=0]
- 16. Protocol for Mesenchymal Stem Cell Isolation [https://fujifilmbiosciences.fujifilm.com/protocol-for-mesenchymal-stem-cell-isolation]
- 17. Mesenchymal stromal cell isolation from pond slider ... [https://www.frontiersin.org/journals/veterinary-science/articles/10.3389/fvets.2025.1546091/full]
- 18. Extracellular matrix and proteolysis: mechanisms driving ... [https://pubmed.ncbi.nlm.nih.gov/41178498/]
- 19. The pros and cons of mechanical dissociation ... [https://www.sciopen.com/article/10.26599/CO.2024.9410009]
- 20. Unlocking the Potential of Collagenases: Structures ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11458858/]
- 21. Tissue Dissociation Guide: Collagenase, Dispase, and ... [https://www.sigmaaldrich.com/US/en/technical-documents/technical-article/research-and-disease-areas/cell-signaling/collagenase-guide?srsltid=AfmBOopnDFElnL8FrsZQ-MtBCRYYymdvLkzOaLCzCkGL1Lxxqj4npt0D]
- 22. Collagenase Wiki - Overview of types and their applications [https://cellsystems.eu/wiki/collagenase-wiki/]
- 23. Trypsin [https://en.wikipedia.org/wiki/Trypsin]
- 24. Comparison of the enzymatic efficiency of Liberase TM ... - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC5985056/]
- 25. Comparative analysis of human Mesenchymal Stromal Cells ... [https://biologydirect.biomedcentral.com/articles/10.1186/s13062-025-00697-w]
- 26. Human Mesenchymal Stem Cells & Growth Media [https://www.merckmillipore.com/HN/es/technical-documents/protocol/cell-culture-and-cell-culture-analysis/primary-cell-culture/hematopoietic-stem-cell-media/mesenchymal-stem-cell-culture]
- 27. A robust and standardized method to isolate and expand ... [https://www.sciencedirect.com/science/article/pii/S1465324923010071]
- 28. Validated methods for isolation and qualification of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12403454/]
- 29. Wharton's Jelly Mesenchymal Stem Cell: Various Protocols ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6885715/]
- 30. A GMP-compliant manufacturing method for Wharton's jelly ... [https://stemcellres.biomedcentral.com/articles/10.1186/s13287-024-03725-0]
- 31. A Simple High Yield Technique for Isolation of Wharton's ... [https://www.ajmb.org/Article?id=60572]
- 32. Evaluation of a new standardized enzymatic isolation ... [https://www.sciencedirect.com/science/article/abs/pii/S0887233314002483]
- 33. Optimization of human umbilical cord mesenchymal stem ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3967601/]
- 34. A Simple High Yield Technique for Isolation of Wharton's ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11007369/]
- 35. Isolation and Culture of Primary Human Dental Pulp Cells ... [https://www.mdpi.com/2409-9279/7/2/22]
- 36. Methodological Stumbles in the Culture of Stem Cells from ... [https://opendentistryjournal.com/VOLUME/19/ELOCATOR/e18742106361595/FULLTEXT/]
- 37. Dental pulp stem cells isolation [https://bio-protocol.org/exchange/minidetail?id=6687148&type=30]
- 38. EP4.8 Isolation of Macrophages and Fibroblasts during Hip ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11954084/]
- 39. Molecular maps of synovial cells in inflammatory arthritis ... [https://www.sciencedirect.com/science/article/pii/S2589004224009295]
- 40. Protocol for the generation and analysis of organoids ... [https://www.sciencedirect.com/science/article/abs/pii/S0040816625002903]
- 41. Comparison between manual and automated methods for ... [https://www.frontiersin.org/journals/cell-and-developmental-biology/articles/10.3389/fcell.2025.1556697/full]
- 42. Characterisation of Mesenchymal Stromal Cells (MSCs ... [https://www.mdpi.com/2077-0383/14/10/3474]
- 43. Comparative analysis of human Mesenchymal Stromal ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12616983/]
- 44. Good Manufacturing Practice for ATMPs [https://www.eurogct.org/research-pathways/manufacturing/good-manufacturing-practice-atmps]
- 45. GMP guidelines for ATMP manufacturing: A practical guide to ... [https://www.crbgroup.com/insights/biotechnology/gmp-guidelines-for-atmp-manufacturing]
- 46. EMA Proposes Revisions to GMP Guideline for ATMPs [https://www.gmp-compliance.org/gmp-news/ema-proposes-revisions-to-gmp-guideline-for-atmps]
- 47. Proposed Revision of EU GMP Part IV, ATMPs [https://www.nsf.org/life-science-regulatory-news/proposed-revision-of-eu-gmp-part-iv-atmps]
- 48. Comparing Mesenchymal Stem Cell (MSC) Isolation ... [https://www.corning.com/worldwide/en/products/life-sciences/resources/stories/the-cutting-edge/comparing-MSC-isolation-techniques.html]
- 49. Optimization of Mesenchymal Stromal Cell (MSC ... [https://www.frontiersin.org/journals/immunology/articles/10.3389/fimmu.2022.918565/full]
- 50. A GMP-compliant manufacturing method for Wharton's jelly ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11069138/]
- 51. Optimizing mesenchymal stem cell therapy: from isolation ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12054297/]
- 52. Isolation of equine multipotent mesenchymal stromal cells by ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4228449/]
- 53. Therapeutic potential of mesenchymal stem cell-based ... [https://www.sciencedirect.com/science/article/pii/S0006291X2501678X?dgcid=rss_sd_all]
- 54. A comprehensive summary of the GISM annual meeting 2025 [https://pmc.ncbi.nlm.nih.gov/articles/PMC12540258/]
- 55. Mechanical isolation of stromal vascular fraction from adipose ... [https://stemcellres.biomedcentral.com/articles/10.1186/s13287-025-04641-7]
- 56. Optimization of Culture Media for Human Umbilical Cord- ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12638159/]
- 57. A New Predictive Technology for Perinatal Stem Cell ... [https://www.mdpi.com/2072-666x/12/7/782]
- 58. Phenotypic and Functional Characterization of Bovine ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11083126/]
- 59. Liberase - an overview | ScienceDirect Topics [https://www.sciencedirect.com/topics/medicine-and-dentistry/liberase]
- 60. Donor age and breed determine mesenchymal stromal cell ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11871689/]
- 61. Development of an effective dissociation protocol for ... [https://www.e-jarb.org/journal/view.html?doi=10.12750/JARB.38.1.10]
- 62. In Vitro Study of a Novel Vibrio alginolyticus-Based ... [https://www.mdpi.com/2073-4409/12/16/2025]
- 63. Comparison of 3D culture systems on mesenchymal stem cell ... [https://pubmed.ncbi.nlm.nih.gov/41077855/]
- 64. Manufacturing mesenchymal stromal cells in a microcarrier ... [https://translational-medicine.biomedcentral.com/articles/10.1186/s12967-024-05373-7]
- 65. Comparison of umbilical cord tissue-derived mesenchymal ... [https://www.sciencedirect.com/science/article/pii/S1465324920307593]
- 66. MSC Renamed? From "Stem Cells" to "Stromal Cells" [https://yoconcelltherapy.com/msc-renamed-from-stem-cells-to-stromal-cells-three-major-shifts-to-note/]
- 67. of equine multipotent Isolation by... mesenchymal stromal cells [https://bmcvetres.biomedcentral.com/articles/10.1186/1746-6148-9-221]
- 68. Clinical Protocols for the Isolation and Expansion of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3076361/]
- 69. Validation of a color deconvolution method to quantify MSC ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9608146/]
- 70. STEMdiff™ Trilineage Differentiation Assay Kit [https://www.stemcell.com/products/stemdiff-trilineage-differentiation-kit.html]
- 71. Mesenchymal Stromal/Stem Cells Isolated by Explant ... [https://www.mdpi.com/2076-3417/14/17/8036]
- 72. Mesenchymal stem cells in treating human diseases [https://www.nature.com/articles/s41392-025-02313-9]
- 73. Transcriptomic Heterogeneity of Human Mesenchymal Stem Cells Derived from Bone Marrow, Dental Pulp, Adipose Tissue, and Umbilical Cord - PubMed [https://pubmed.ncbi.nlm.nih.gov/37384924/]
- 74. Comparison of the therapeutic effects of mesenchymal stem cells derived from human dental pulp (DP), adipose tissue (AD), placental amniotic membrane (PM), and umbilical cord (UC) on postmenopausal osteoporosis - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC11004311/]
- 75. Adipose derived stem cells – Sources, differentiation ... [https://www.sciencedirect.com/science/article/pii/S0753332225002306]
- 76. Frontiers | Comparison of the therapeutic effects of mesenchymal stem cells derived from human dental pulp (DP), adipose tissue (AD), placental amniotic membrane (PM), and umbilical cord (UC) on postmenopausal osteoporosis [https://www.frontiersin.org/journals/pharmacology/articles/10.3389/fphar.2024.1349199/full]
- 77. An enzyme-free technique enables the isolation of a large ... [https://www.nature.com/articles/s41598-023-34915-0]
- 78. Size-dependent immunomodulation by MSC secretome [https://pmc.ncbi.nlm.nih.gov/articles/PMC12512777/]
- 79. Fine-tuning licensing strategies to boost MSC-based ... [https://stemcellres.biomedcentral.com/articles/10.1186/s13287-025-04315-4]
- 80. Mesenchymal Stromal Cell-Based Products: Challenges ... [https://www.mdpi.com/1422-0067/25/11/6063]