Osteogenic, Chondrogenic, and Adipogenic Differentiation of Stem Cells: Mechanisms, Methods, and Clinical Translation
This comprehensive review synthesizes current advancements in steering mesenchymal stem cell (MSC) differentiation towards osteogenic, chondrogenic, and adipogenic lineages.
Osteogenic, Chondrogenic, and Adipogenic Differentiation of Stem Cells: Mechanisms, Methods, and Clinical Translation
Abstract
This comprehensive review synthesizes current advancements in steering mesenchymal stem cell (MSC) differentiation towards osteogenic, chondrogenic, and adipogenic lineages. It explores the foundational biology, including key transcription factors and signaling pathways, and details innovative methodological approaches such as 3D bioprinting, advanced biomaterials, and machine learning for predicting differentiation. The article also addresses critical challenges in optimization and standardization, provides a comparative analysis of MSC sources, and discusses validation strategies for clinical application. Aimed at researchers, scientists, and drug development professionals, this resource bridges fundamental research with translational medicine, offering insights for regenerative therapies in orthopedics and beyond.
The Biological Blueprint: Unraveling Lineage-Specific Pathways and Transcriptional Networks
Mesenchymal stem cells (MSCs) have emerged as a highly promising strategy in regenerative medicine due to their self-renewal, pluripotency, and immunomodulatory properties [1]. These non-hematopoietic, multipotent stem cells were first identified in bone marrow and can differentiate into various mesodermal lineages while modulating the immune system [1]. The therapeutic potential of MSCs from different tissues has been widely explored in preclinical models and clinical trials for human diseases, ranging from autoimmune and inflammatory disorders to neurodegenerative diseases and orthopedic injuries [1]. According to the International Society for Cellular Therapy (ISCT), MSCs are defined by three key criteria: adherence to plastic under standard culture conditions; expression of specific surface markers (CD73, CD90, and CD105 ≥95%) while lacking hematopoietic markers (CD34, CD45, CD14/CD11b, CD79α/CD19, HLA-DR ≤2%); and capacity to differentiate into osteogenic, chondrogenic, and adipogenic lineages in vitro [1] [2]. Originally termed "mesenchymal stem cells" by Dr. Arnold Caplan in 1991, the nomenclature has evolved, with the ISCT now officially defining them as "Mesenchymal Stromal Cells" to reflect their tissue-supporting and immunomodulatory functions [3] [4].
Defining Characteristics: Surface Markers and Identification
The precise identification of MSCs relies on a specific immunophenotypic profile established by international standards. The positive and negative marker expression provides a critical framework for researchers to validate MSC populations before experimental or therapeutic application.
Table 1: Essential Surface Markers for MSC Identification According to ISCT Criteria
| Marker Category | Specific Markers | Expression Requirement | Biological Significance |
|---|---|---|---|
| Positive Markers | CD105 (Endoglin) | ≥95% expression | Type I membrane glycoprotein essential for cell migration and angiogenesis [1]. |
| CD73 (5'-ectonucleotidase) | ≥95% expression | Catalyzes AMP hydrolysis to adenosine; role in cell signaling within bone marrow [1]. | |
| CD90 (Thy-1) | ≥95% expression | GPI-anchored protein mediating cell-cell and cell-ECM interactions; contributes to adhesion and migration [1]. | |
| Negative Markers | CD45, CD34 | ≤2% expression | CD45: marker for white blood cells; CD34: biomarker for hematopoietic stem cells [1]. |
| CD14/CD11b | ≤2% expression | Expressed on monocytes and macrophages [1]. | |
| CD79α/CD19 | ≤2% expression | Markers of B cells [1]. | |
| HLA-DR | ≤2% expression | MHC class II molecule with strong immunogenic properties [1]. |
The adherence to these marker criteria is crucial for ensuring population purity and distinguishing MSCs from hematopoietic cells. Additional markers like STRO-1, CD146, and CD29 are often used in research to identify subpopulations with enhanced stemness, but the core ISCT panel remains the standard for minimal definition [5]. The expression profile must be confirmed using techniques such as flow cytometry, and researchers should note that marker expression can be influenced by factors like passage number and culture conditions [5].
MSCs can be isolated from a remarkable variety of adult and perinatal tissues, each source offering distinct advantages and challenges. The selection of a source material is a critical first step that influences the yield, proliferation rate, and potential application of the derived MSCs.
Table 2: Comparison of Primary Mesenchymal Stem Cell Sources
| Tissue Source | Isolation Yield & Key Features | Primary Isolation Methods | Research & Clinical Relevance |
|---|---|---|---|
| Bone Marrow (BM-MSCs) | Limited yield (0.01-0.001% of nucleated cells); considered the "gold standard" [2]. | Bone marrow aspirate followed by density gradient centrifugation (e.g., Ficoll-Paque) and adherence culture [3]. | Most extensively studied source; used in 10 approved therapies; requires invasive harvest [1] [2]. |
| Adipose Tissue (AD-MSCs) | High yield (up to 1 billion cells from 300g tissue); less invasive harvest [2]. | Lipoaspirate processed via enzymatic digestion (e.g., collagenase) and red blood cell lysis [3] [4]. | Abundant source; advantages in bone regeneration and skin healing; three approved therapies [2]. |
| Umbilical Cord (UC-MSCs) | High concentration in Wharton's Jelly; enhanced proliferation, low immunogenicity [1] [2]. | Enzymatic digestion of cord tissue or explant culture; ISO/TS 22859-1:2022 standard exists [2] [3]. | Ideal for allogeneic transplantation; three approved therapies [2]. |
| Umbilical Cord Blood (UCB-MSCs) | Contains MSCs alongside hematopoietic stem cells; lower yield than UC [2]. | Density gradient centrifugation of cord blood to isolate mononuclear cells [2] [3]. | High proliferation and clonogenic rates; delayed senescence [2]. |
| Placenta (P-MSCs) | Complex organ with high MSC concentration; superior proliferative capacity [2]. | Surgical dissection of specific regions (amnion, chorion) followed by enzymatic digestion [3]. | Exhibits pronounced immunosuppressive effects; isolation challenged by complex composition [2]. |
| Menstrual Blood/Endometrium (MenSCs/eMSCs) | Easy, non-invasive collection; rapid doubling time (~20 hours) [2]. | Collection and processing of menstrual effluent or endometrial biopsy via adherence [2]. | Promising for gynecological applications; no clinical trials to date [2]. |
Detailed Protocol: Isolation of Human Umbilical Cord MSCs (Wharton's Jelly Derivation)
Principle: This protocol utilizes enzymatic digestion to efficiently release MSCs from the Wharton's Jelly matrix of the human umbilical cord, providing a high yield of cells suitable for allogeneic therapies [2] [3].
Reagents and Materials:
- Fresh human umbilical cord (full-term, maternal consent obtained)
- Dulbecco's Phosphate Buffered Saline (DPBS), sterile
- Antibiotic-Antimycotic solution (100X)
- Collagenase Type I or Type IV (e.g., 1-2 mg/mL working concentration)
- Hyaluronidase (optional, to enhance digestion)
- Fetal Bovine Serum (FBS) to quench digestion
- Complete culture medium: α-MEM or DMEM/F12, 10% FBS, 1% Antibiotic-Antimycotic
- Tissue culture plasticware (flasks, plates)
- Surgical scissors, forceps, scalpels
Procedure:
- Transport and Washing: Transport the umbilical cord to the lab in a sterile container with cold DPBS containing 1% Antibiotic-Antimycotic. Wash thoroughly in fresh DPBS to remove residual blood.
- Vessel Removal: Using sterile instruments, dissect the cord to remove the two arteries and one vein.
- Tissue Mincing: Chop the remaining Wharton's Jelly tissue into small fragments (~1-2 mm³).
- Enzymatic Digestion: Incubate the tissue fragments in a collagenase solution (1-2 mg/mL in DPBS) for 3-6 hours at 37°C with gentle agitation.
- Digestion Quenching: Add an equal volume of complete culture medium containing 10% FBS to stop the enzymatic reaction.
- Filtration and Centrifugation: Filter the cell suspension through a 100μm cell strainer to remove undigested tissue. Centrifuge the filtrate at 300-400 x g for 10 minutes.
- Plating and Culture: Resuspend the cell pellet in complete culture medium and plate in a tissue culture flask. Maintain at 37°C in a 5% CO₂ humidified incubator.
- Medium Changes: Perform the first medium change after 48-72 hours to remove non-adherent cells, then change the medium every 3-4 days thereafter.
- Passaging: Passage cells at 70-80% confluence using standard trypsin/EDTA digestion.
Notes: The isolated cells should be characterized according to ISCT criteria (Section 2) before experimental use. The international standard ISO/TS 22859-1:2022 provides further technical specifications for hUC-MSCs [2].
The Scientist's Toolkit: Essential Research Reagent Solutions
Table 3: Key Reagents for MSC Research and Their Applications
| Reagent / Material | Function in MSC Research | Example Application & Notes |
|---|---|---|
| Fetal Bovine Serum (FBS) | Provides essential growth factors, hormones, and nutrients for in vitro cell growth. | Standard component (typically 10-20%) of basal MSC culture medium. Batch variability is a significant concern; screening is recommended. |
| Collagenase Type I/IV | Enzyme that degrades collagen, a major ECM component, to dissociate cells from tissues. | Critical for isolating MSCs from adipose tissue (lipoaspirate) and umbilical cord [3]. |
| Ficoll-Paque Premium | Density gradient medium for isolating mononuclear cells from bone marrow or cord blood. | Enriches for MSCs by separating them from red blood cells and granulocytes [3]. |
| Trypsin-EDTA Solution | Proteolytic enzyme (Trypsin) chelates calcium (EDTA) to dissociate adherent cells for passaging. | Standard reagent for detaching adherent MSCs from culture plastic for subculturing. |
| CD105, CD73, CD90 Antibodies | Conjugated antibodies for flow cytometry to confirm the positive marker profile of MSCs. | Required for immunophenotypic characterization per ISCT guidelines. |
| Osteo-/Chondro-/Adipo-Induction Media | Specialized media containing specific inducing factors to drive MSC differentiation. | Used to validate MSC multipotency in vitro (see Section 5). |
| Acrylate-based Functionalized Substrates | Synthetic polymer supports grafted with biomolecules (gelatin, heparin) to study cell-biomaterial interactions. | Used in advanced research to direct MSC fate and for bone tissue engineering applications [6]. |
| CRISPR-Cas9 System | Gene editing tool to create specific genetic modifications in MSCs for functional studies. | Used to investigate gene function, e.g., role of XYLT1 in chondrogenic differentiation [7]. |
Multipotency: Trilineage Differentiation Potential
The defining functional characteristic of MSCs is their ability to differentiate into osteoblasts, chondrocytes, and adipocytes in vitro. This trilineage potential is not only a validation criterion but also the foundation of their application in regenerative medicine. The differentiation processes are governed by complex and highly regulated transcriptional networks [5].
Detailed Protocol: Standard In Vitro Trilineage Differentiation Assay
Principle: This protocol provides a standardized method to induce and validate the adipogenic, osteogenic, and chondrogenic differentiation of MSCs in vitro, fulfilling a core defining criterion [1] [5].
Reagents and Materials:
- Validated MSCs (Passage 2-4, ~70-80% confluence)
- Basal medium: DMEM high glucose, 10% FBS, 1% Penicillin/Streptomycin
- Adipogenic Induction Medium: Basal medium supplemented with 0.5 mM IBMX, 1 μM Dexamethasone, 10 μM Insulin, 200 μM Indomethacin.
- Adipogenic Maintenance Medium: Basal medium supplemented with 10 μM Insulin.
- Osteogenic Induction Medium: Basal medium supplemented with 0.1 μM Dexamethasone, 10 mM β-Glycerophosphate, 50 μM Ascorbic Acid-2-Phosphate.
- Chondrogenic Induction Medium: Serum-free DMEM high glucose supplemented with 1% ITS+ Premix, 0.1 μM Dexamethasone, 50 μM Ascorbic Acid-2-Phosphate, 40 μg/mL Proline, and 10 ng/mL TGF-β3.
- Oil Red O staining solution
- Alizarin Red S staining solution
- Alcian Blue staining solution
Procedure:
- Cell Seeding:
- Adipogenesis/Osteogenesis: Seed MSCs in 12- or 24-well plates at a standard density (e.g., 2.1 x 10⁴ cells/cm² for adipogenesis, 3.1 x 10⁴ cells/cm² for osteogenesis). Allow cells to adhere for 24 hours.
- Chondrogenesis: Pellet 2.5 x 10⁵ MSCs in a 15 mL polypropylene tube by centrifugation (500 x g for 5 minutes). Culture the pellet in the tube.
Induction:
- Adipogenesis: Once cells are 100% confluent, replace the basal medium with Adipogenic Induction Medium. After 3 days, switch to Adipogenic Maintenance Medium for 24 hours. Repeat this cycle 3-4 times. Finally, maintain cells in Maintenance Medium for an additional 4-7 days, changing medium every 2-3 days.
- Osteogenesis: Once cells are 70-80% confluent, replace the basal medium with Osteogenic Induction Medium. Maintain for 21 days, changing the medium every 3-4 days.
- Chondrogenesis: Loosen the cell pellet by gently tapping the tube and add Chondrogenic Induction Medium. Incubate for 21-28 days, changing the medium every 2-3 days. The pellet will form a spherical micromass.
Staining and Analysis:
- Adipogenesis: Fix differentiated cells with 4% PFA for 20 minutes. Stain with Oil Red O (working solution) for 30-60 minutes to visualize intracellular lipid droplets. Quantify by eluting the stain with isopropanol and measuring absorbance at 520 nm.
- Osteogenesis: Fix cells with 4% PFA for 20 minutes. Stain with 2% Alizarin Red S (pH 4.1-4.3) for 20-30 minutes to detect calcium deposits.
- Chondrogenesis: Fix the micromass pellet with 4% PFA, process for paraffin embedding, and section. Stain sections with Alcian Blue (pH 2.5) for 30 minutes to visualize sulfated proteoglycans in the extracellular matrix.
Troubleshooting: Lack of differentiation may indicate over-passaged MSCs, suboptimal inducer concentrations, or poor initial cell quality. Always include undifferentiated controls (cultured in basal medium) for comparison. For molecular validation, perform RT-qPCR for lineage-specific markers (Figure 2).
Clinical Applications and Future Perspectives
The therapeutic application of MSCs has expanded dramatically, with over ten approved MSC-based therapies marketed worldwide and hundreds of clinical trials ongoing [2] [8]. The initial focus on their regenerative potential via direct differentiation has shifted towards appreciating their potent paracrine effects. MSCs release a diverse array of bioactive molecules, including growth factors, cytokines, and extracellular vesicles (EVs), which play crucial roles in modulating the local cellular environment, promoting tissue repair, angiogenesis, and exerting anti-inflammatory and immunomodulatory effects [1] [8] [4].
Approved MSC therapies primarily address conditions like complex perianal fistulas in Crohn's disease, graft-versus-host disease (GVHD), and amyotrophic lateral sclerosis [8]. In gynecology, MSC therapies for uterine adhesions and early-onset ovarian failure have progressed to clinical application, demonstrating notable efficacy [2]. However, the field faces challenges, including inconsistent efficacy in clinical trials, product heterogeneity, and a lack of standardized manufacturing and delivery protocols [8] [4]. Future directions involve overcoming these hurdles through strategies like genetic modification, preconditioning ("priming") of MSCs, and a growing interest in cell-free therapies utilizing MSC-derived extracellular vesicles [4]. A deeper understanding of MSC biology, differentiation pathways, and mechanisms of action will undoubtedly pave the way for more effective and reliable regenerative therapies.
Osteogenic differentiation is a sophisticated, multi-step process through which mesenchymal stem cells (MSCs) commit to the osteoblast lineage, ultimately producing bone-forming cells responsible for bone matrix synthesis and mineralization. This process is governed by a precise transcriptional hierarchy and influenced by several key signaling pathways. Understanding this regulatory network is paramount for advancing bone tissue engineering, regenerative medicine, and developing therapeutics for bone loss diseases such as osteoporosis. The master transcription factor Runt-related transcription factor 2 (Runx2) initiates the osteogenic program, while the zinc-finger transcription factor Osterix (Osx) acts downstream as an essential regulator for osteoblast maturation and bone matrix deposition [9] [10]. The differentiation process is further fine-tuned by major signaling pathways, notably the Wnt/β-catenin and Bone Morphogenetic Protein (BMP)/Smad pathways, which integrate external cues to regulate the activity of these core transcription factors [11] [12] [13]. This application note provides a detailed overview of these regulators, their functional crosstalk, and standard experimental protocols for investigating osteogenic differentiation in vitro.
Master Transcriptional Regulators of Osteogenesis
Runx2: The Master Switch of Osteoblast Lineage
Runx2 is a transcription factor belonging to the runt homology domain protein family and is widely recognized as the master regulator of osteoblast differentiation [9] [14].
- Function and Expression: Runx2 is expressed early in cells prefiguring the skeleton and is essential for directing MSCs into the osteoblast lineage. It controls the expression of major bone matrix protein genes by binding to the osteoblast-specific cis-acting element (OSE2) in the promoters of genes such as osteocalcin (OC), osteopontin (OPN), bone sialoprotein (BSP), and collagen type I alpha 1 (Col1A1) [9]. Its expression and transcriptional activity must be precisely regulated, as both deficiency and overexpression lead to bone defects. Homozygous loss of Runx2 in mice results in a complete lack of bone formation due to maturational arrest of osteoblasts [9]. In humans, haploinsufficiency of RUNX2 causes cleidocranial dysplasia (CCD), characterized by hypoplastic/aplastic clavicles and open fontanelles [9] [14] [10].
- Regulation by Post-Translational Modifications (PTMs): Runx2 activity is intricately controlled by PTMs, including phosphorylation, prolyl isomerization, acetylation, and ubiquitination [14]. For instance, FGF signaling activates ERK and p38 MAPKs, which phosphorylate Runx2. This phosphorylation facilitates prolyl isomerization by PIN1, which in turn exposes lysine residues for acetylation by p300. Acetylation stabilizes Runx2 by protecting it from ubiquitin-mediated degradation, thereby enhancing its transactivation potential [14].
Osterix (Osx): The Gatekeeper of Osteoblast Maturation
Osterix (Osx or Sp7) is a zinc finger-containing transcription factor that acts downstream of Runx2 and is indispensable for osteoblast maturation [9] [10].
- Function and Expression: Mice lacking Osx completely fail to form bone and are devoid of mature osteoblasts, demonstrating its non-redundant role [9] [10]. Osx is required for the differentiation of preosteoblasts into fully functional osteoblasts and osteocytes. It regulates a suite of genes involved in the final stages of osteogenic differentiation and bone matrix production.
- Interaction with Runx2 and PPARγ: Osx and Runx2 can functionally interact to cooperatively induce the expression of osteogenic genes like Col1a1 and Ibsp. This cooperation is mediated by their binding to adjacent Sp1 and Runx sites on target gene enhancers and is regulated by MAPK signaling [15]. Beyond its osteogenic role, Osx also represses adipogenesis by directly interacting with the ligand-binding domain of PPARγ, the master regulator of adipogenesis. This interaction inhibits PPARγ's transcriptional activity, thereby suppressing fat cell differentiation and promoting a commitment to the osteoblastic lineage [16].
Table 1: Key Master Transcription Factors in Osteogenic Differentiation
| Transcription Factor | Key Function | Genetic Evidence (Loss-of-Function) | Key Downstream Targets |
|---|---|---|---|
| Runx2 | Master regulator; initiates osteoblast lineage commitment from MSCs. | Complete lack of bone formation; arrested osteoblast maturation [9]. | Osteocalcin (OC), Osteopontin (OPN), Bone Sialoprotein (BSP), Collagen type I (Col1A1) [9]. |
| Osterix (Osx) | Essential for osteoblast maturation and bone matrix deposition. | No bone formation; complete absence of mature osteoblasts [9] [10]. | A repertoire of genes for osteoblast maturation and matrix mineralization [10]. |
Key Signaling Pathways in Osteogenic Differentiation
Wnt/β-catenin Signaling Pathway
The canonical Wnt/β-catenin pathway is a critical regulator of bone mass and osteoblastogenesis [11] [12] [17].
- Mechanism: In the absence of a Wnt ligand, a cytoplasmic "destruction complex" (containing Axin, APC, GSK-3β, and CK1) phosphorylates β-catenin, targeting it for proteasomal degradation. When Wnt ligands (e.g., Wnt1, Wnt3a) bind to the Frizzled (Fzd) receptor and LRP5/6 co-receptor, this complex is disrupted. This leads to the stabilization and accumulation of β-catenin in the cytoplasm, followed by its translocation to the nucleus. Inside the nucleus, β-catenin partners with TCF/LEF transcription factors to activate the expression of target genes, including those critical for osteoblast differentiation and proliferation [11] [17].
- Role in Osteogenesis: Wnt/β-catenin signaling promotes MSC commitment to the osteoblast lineage and simultaneously inhibits differentiation into the adipocyte and chondrocyte lineages [12]. Activation of this pathway results in increased bone mass and bone formation strength in vivo [12].
BMP/Smad Signaling Pathway
The BMP pathway is a potent osteoinductive signal that works in concert with other pathways to drive bone formation [12] [13].
- Mechanism: BMP ligands (e.g., BMP2, BMP4, BMP7) bind to a heterodimeric complex of type I and type II BMP transmembrane serine/threonine kinase receptors. This binding leads to the phosphorylation of receptor-regulated Smads (R-Smads: Smad1, Smad5, Smad8). The phosphorylated R-Smads then form a complex with the common mediator Smad4 (Co-Smad). This trimeric complex translocates into the nucleus, where it regulates the transcription of target genes, including Runx2 [12] [13].
- Role in Osteogenesis: BMP signaling is a key upstream activator of the osteogenic transcription factor cascade. It stimulates the expression of Runx2, which in turn drives the entire osteogenic program. The pathway is tightly controlled by inhibitory Smads (I-Smads), such as Smad6 and Smad7, which provide a negative feedback loop to prevent over-signaling [13].
Crosstalk Between Signaling Pathways
A critical aspect of osteogenic control is the crosstalk between different signaling pathways. The Wnt/β-catenin and BMP/Smad pathways, while distinct, do not operate in isolation. They exhibit significant functional synergy to promote robust osteogenic differentiation [12]. Active Wnt/β-catenin signaling can promote the expression of downstream targets of the BMP signaling pathway, creating a reinforced pro-osteogenic network. This integration ensures that MSCs receive coordinated signals to commit to the bone lineage effectively.
Experimental Protocols for Studying OsteogenesisIn Vitro
Standard Osteogenic Differentiation of Mesenchymal Stem Cells (MSCs)
This protocol outlines the basic methodology for inducing and assessing osteogenic differentiation in MSCs in vitro.
- Cell Culture: Seed human bone marrow-derived MSCs (BMSCs) or an appropriate MSC line at a density of 5,000 - 10,000 cells/cm² in basal growth medium (e.g., α-MEM or DMEM) supplemented with 10% Fetal Bovine Serum (FBS) and 1% penicillin/streptomycin.
- Osteogenic Induction: Once cells reach 80-90% confluence, replace the growth medium with osteogenic induction medium. The basal medium is supplemented with:
- 50-100 µM Ascorbic Acid-2-phosphate: Promotes collagen matrix synthesis.
- 10 mM β-glycerophosphate: Provides a source of organic phosphate for matrix mineralization.
- 10-100 nM Dexamethasone: A synthetic glucocorticoid that promotes osteoblast differentiation by modulating transcription factors like Runx2 [9].
- Maintenance: Culture the cells for 21-28 days, changing the osteogenic medium every 2-3 days.
Modulating Signaling Pathways: Application of Recombinant Proteins and Inhibitors
To investigate the role of specific pathways, the osteogenic medium can be supplemented with activating or inhibiting agents.
Table 2: Reagents for Modulating Key Osteogenic Pathways
| Target Pathway | Reagent Example | Concentration Range | Function / Effect |
|---|---|---|---|
| BMP Signaling | Recombinant human BMP-2 | 50-100 ng/mL [18] | Potent osteoinductive factor; activates BMP-Smad signaling to induce Runx2 expression. |
| Wnt/β-catenin Signaling | Recombinant Wnt3a | 10-100 ng/mL | Activates canonical Wnt signaling to promote osteoblast lineage commitment. |
| CHIR99021 (GSK-3β inhibitor) | 3-10 µM | Chemical activator of Wnt signaling by inhibiting β-catenin degradation. | |
| FGF Signaling | Basic FGF (bFGF/FGF-2) | 5-20 ng/mL [18] | Mitogen for MSCs; its effect on osteogenesis is stage-dependent (inhibitory early, promotive later). |
| TGF-β Signaling | Recombinant TGF-β1 | 1-5 ng/mL [18] | Low concentrations (e.g., 1 ng/mL) may promote osteogenesis, while high concentrations inhibit it. |
Assessment of Osteogenic Differentiation
- Alkaline Phosphatase (ALP) Activity: Measure ALP activity (an early osteoblast marker) around day 7-10 using a colorimetric assay (e.g., with p-nitrophenyl phosphate) and normalize to total protein content.
- Matrix Mineralization (Alizarin Red S Staining): After 21-28 days, fix cells and stain with 2% Alizarin Red S (pH 4.2) to detect calcium deposits. Stained mineralized nodules can be quantified by eluting the dye and measuring absorbance or by image analysis.
- Gene Expression Analysis (qRT-PCR): Isolate RNA at specific time points (e.g., days 7, 14, 21). Analyze the expression of osteogenic markers:
- Early: Runx2, ALP
- Mid/Late: Osterix (Sp7), Osteopontin (SPP1), Bone Sialoprotein (BSP)
- Late: Osteocalcin (OC)
- Protein Analysis (Western Blot/Immunofluorescence): Confirm the expression and localization of key proteins like Runx2, Osterix, and phosphorylated Smads (p-Smad1/5/8).
The Scientist's Toolkit: Key Research Reagent Solutions
Table 3: Essential Reagents for Osteogenesis Research
| Reagent / Material | Function / Application | Example & Notes |
|---|---|---|
| Mesenchymal Stem Cells (MSCs) | Primary model system for in vitro osteogenesis. | Human Bone Marrow MSCs (hBM-MSCs), Adipose-derived MSCs (ADSCs), or cell lines like C3H10T1/2 or MC3T3-E1. |
| Osteogenic Induction Supplements | Core components to induce differentiation in basal medium. | Ascorbic Acid, β-Glycerophosphate, and Dexamethasone. Available as pre-mixed supplements from various suppliers. |
| Recombinant Growth Factors | To activate specific pro-osteogenic signaling pathways. | Recombinant Human BMP-2, Wnt3a, FGF-2, TGF-β1. Use at optimized concentrations to avoid off-target effects. |
| Small Molecule Inhibitors/Activators | To chemically perturb pathways and study their function. | CHIR99021 (Wnt activator), SB431542 (TGF-β inhibitor), Dorsomorphin (BMP inhibitor). |
| Histological Stains | To detect and quantify terminal differentiation markers. | Alizarin Red S (mineralization), Oil Red O (lipid droplets, for adipogenesis control), Von Kossa (calcium phosphate). |
| Antibodies | For protein-level analysis of transcription factors and signaling molecules. | Anti-Runx2, Anti-Osterix, Anti-p-Smad1/5/8, Anti-β-catenin. Validate for application (WB, IF). |
Integrated Osteogenic Regulatory Network
The following diagram synthesizes the key regulators and their interactions described in this note into a cohesive osteogenic differentiation network.
Chondrogenic differentiation, the process by which progenitor cells differentiate into chondrocytes to form cartilage, is a critical pathway in skeletal development, adult homeostasis, and tissue regeneration. This process is tightly regulated by a complex interplay of transcription factors and extracellular cues from the cellular microenvironment, or niche. The transcription factor SRY-box transcription factor 9 (Sox9) is the undisputed master regulator of this pathway, essential for directing mesenchymal progenitor cells toward a chondrogenic fate. Its activity, in combination with other SOX proteins and in response to specific signaling pathways, orchestrates the expression of key cartilage-specific extracellular matrix (ECM) components. Beyond intracellular regulators, the cartilage niche—comprising the native cellular environment, structural components, and physicochemical signals—plays a decisive role in determining the ultimate phenotype and stability of the differentiated cartilage. This application note details the core mechanisms of Sox9 action and the influential role of the niche, providing structured data and validated protocols to support research and development in cartilage biology and regenerative medicine.
Key Players & Molecular Mechanisms
The Sox Trio and Transcriptional Regulation
The core transcriptional machinery driving chondrogenesis is the Sox Trio, consisting of Sox9, L-Sox5 (Sox5), and Sox6. These factors cooperate to activate the gene expression program essential for chondrocyte differentiation and cartilage matrix synthesis [19] [20].
- Sox9: Functions as the master transcription factor. It binds to the consensus DNA sequence (A/T)(A/T)CAA(A/T)G in the enhancer regions of its target genes. It is indispensable for the initiation of chondrogenesis and for the expression of key ECM genes like type II collagen (COL2A1) and aggrecan (ACAN) [19] [20]. Mutations in SOX9 cause campomelic dysplasia, a severe skeletal dysplasia syndrome, underscoring its non-redundant role [19].
- L-Sox5 and Sox6: These structurally related proteins lack a transactivation domain but potentiate the function of Sox9. They bind to adjacent DNA sites, facilitating the assembly of a more robust enhanceosome complex that dramatically boosts the transcription of cartilage-specific genes [20].
Sox9's activity is itself regulated by multiple mechanisms, including phosphorylation and nuclear translocation, interaction with co-activators like CREB-binding protein (CBP)/p300, and modulation by signaling pathways such as BMP/TGF-β via Smad proteins [19] [20].
Biphasic Expression and Novel Functions of Sox9
Recent research has revealed that Sox9 expression during chondrogenic differentiation is biphasic [19]. An immediate, transient early phase is followed by a later, sustained phase associated with active ECM synthesis. While the late phase is linked to canonical matrix production, the early phase is crucial for preparing the cell for the demanding differentiative process. A seminal study identified a novel essential function for Sox9 during this early phase: the regulation of translational capacity [19].
Early Sox9 knockdown was shown to:
- Severely inhibit chondrogenic differentiation weeks later.
- Downregulate the expression of ribosome biogenesis factors and ribosomal protein subunits.
- Decrease the cell's overall translational capacity, correlating with lower amounts of active mono- and polysomes.
- Alter the mode of translation initiation (cap- vs. IRES-mediated) [19].
This demonstrates that beyond its well-known transcriptional role, Sox9 primes the cellular machinery for the high levels of protein synthesis required for subsequent proliferation and massive ECM production.
The Deterministic Role of the Cartilage Niche
The local microenvironment, or niche, is a dominant factor in specifying the type of cartilage regenerated by stem cells. A systematic in vivo study demonstrated that the native cartilage niche overrides instructively biomimetic scaffolds and co-cultured chondrocytes to determine the final cartilage phenotype [21].
- Key Finding: When BMSCs were implanted into specific native cartilage niches (ear or articular), the type of cartilage regenerated was always consistent with the implantation site, regardless of the type of acellular cartilage sheet (ACS) scaffold or the source of co-cultured chondrocytes used [21].
- Niche-Specific Outcomes:
- The articular cartilage niche regulated BMSCs to regenerate hyaline-like cartilage.
- The ear cartilage niche regulated BMSCs to regenerate elastic cartilage, characterized by elastin expression [21].
This work provides compelling evidence that for clinical translation, strategies must not only focus on inducing chondrogenesis but also on recapitulating or harnessing niche-specific signals to achieve a functionally appropriate and stable cartilage type.
Summarized Quantitative Data
| Experimental Manipulation | Time of Analysis | Key Quantitative Findings & Impact on Chondrogenesis |
|---|---|---|
| Sox9 siRNA Knockdown (prior to differentiation) | 2 hours & 7 days | Severe inhibition of late differentiation (weeks later).↓ Expression of ribosome biogenesis factors and ribosomal proteins.↓ Total translational capacity (SuNSET assay).↓ Amount of active mono- and polysomes (polysome profiling).Altered cap- vs. IRES-mediated translation (bicistronic reporter). |
| Sox9 Overexpression (Lentiviral) | Various time points | Reciprocal effects to knockdown; enhanced chondrogenic capacity. |
| Implanted Construct | Native Niche for Implantation | Resulting Cartilage Type Regenerated by BMSCs |
|---|---|---|
| BMSC + Ear ACS (EACS) | Ear Cartilage | Elastic Cartilage |
| BMSC + Articular ACS (AACS) | Articular Cartilage | Hyaline-like Cartilage |
| BMSC + AACS | Ear Cartilage | Elastic Cartilage |
| BMSC + EACS | Articular Cartilage | Hyaline-like Cartilage |
| BMSC + AACS + Articular Chondrocytes | Ear Cartilage | Elastic Cartilage |
| BMSC + EACS + Ear Chondrocytes | Articular Cartilage | Hyaline-like Cartilage |
Application Notes & Protocols
Protocol: Investigating the Early Role of Sox9 via Knockdown in ATDC5 Cells
This protocol is adapted from methods used to elucidate Sox9's novel role in regulating translational capacity [19].
Objective: To ablate early Sox9 expression and analyze its effects on the transcriptome, proteome, and translational machinery during chondrogenic differentiation.
Materials:
- Cell Line: ATDC5 murine progenitor cells.
- Culture Media: DMEM/F12 + 5% FBS + 1% Antibiotic/Antimycotic + 1% NEAA.
- Differentiation Supplements: 10 µg/mL insulin, 10 µg/mL transferrin, 30 nM sodium selenite.
- Sox9 siRNA: Duplex sequence: sense 5'-GACUCACAUCUCUCCUAAUTT-3', antisense 5'-AUUAGGAGAGAUGUGAGUCTT-3'.
- Control siRNA: Scrambled sequence.
- Transfection Reagent: HiPerFECT or equivalent.
Procedure:
- Cell Seeding: Plate ATDC5 cells at a density of 20,000 cells/cm² in standard growth media.
- Transfection: 24 hours after seeding, transfert cells with 100 nM Sox9 siRNA or Control siRNA using the manufacturer's protocol for HiPerFECT.
- Induction of Differentiation: 24 hours post-transfection, initiate chondrogenic differentiation by replacing the media with culture media containing the differentiation supplements (insulin, transferrin, selenite).
- Harvesting: Harvest samples for analysis at critical time points (e.g., 2 hours for early transcriptomic changes, 7 days for proteomic and differentiation markers).
- RNA-seq: Isolate total RNA using TRIzol for transcriptome analysis.
- Label-free Proteomics: Prepare cell lysates for proteomic analysis.
- Functional Assays:
- SuNSET Assay: Incorporate puromycin to measure de novo protein synthesis.
- Polysome Profiling: Lyse cells and separate ribosomal fractions by sucrose density gradient centrifugation to assess ribosome activity.
- Bicistronic Reporter Assay: Transfect a reporter plasmid to determine the mode of translation initiation.
Protocol: Assessing Niche-Directed Chondrogenesis of BMSCs In Vivo
This protocol outlines the approach for demonstrating the deterministic role of the native cartilage niche [21].
Objective: To test whether a specific native cartilage microenvironment can direct BMSCs to regenerate a matching cartilage type, overriding other biomimetic cues.
Materials:
- Cells: GFP-labeled Bone Marrow Stromal Cells (BMSCs), ear chondrocytes (EACs), articular chondrocytes (ARCs).
- Scaffolds: Acellular Cartilage Sheets (ACS) from ear (EACS) and articular (AACS) cartilage.
- Animals: Immunocompetent large animal model (e.g., pig).
Procedure:
- Construct Preparation:
- Prepare two types of ACSs (EACS and AACS) via decellularization of tissue sheets.
- Seed GFP-BMSCs onto the ACSs alone, or in a "sandwich" model with corresponding chondrocytes (e.g., BMSCs + AACS + ARCs).
- Experimental Groups & Implantation:
- Niche-Matched: Implant BMSC-EACS into ear cartilage niche; BMSC-AACS into articular cartilage niche.
- ACS Niche-Mismatched: Implant BMSC-AACS into ear niche; BMSC-EACS into articular niche.
- Biomimetic Niche-Mismatched: Implant BMSC-AACS-ARC into ear niche; BMSC-EACS-EAC into articular niche.
- In Vivo Culture: Allow constructs to develop in vivo for an extended period (e.g., one year).
- Analysis:
- Cell Tracking: Identify the origin of regenerating cells via GFP fluorescence.
- Histology & Immunohistochemistry: Assess cartilage type using specific markers:
- Hyaline Cartilage: Type II Collagen (Col II), Proteoglycans (Safranin O staining).
- Elastic Cartilage: Elastin.
- Articular Surface Marker: PRG4 (Lubricin).
Signaling Pathways and Workflows
Sox9 in Chondrogenic Differentiation
Experimental Workflow for Niche Studies
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Reagents for Chondrogenesis and Niche Research
| Item | Function/Application | Example from Literature |
|---|---|---|
| ATDC5 Progenitor Cell Line | A well-established in vitro model for studying the stepwise process of chondrogenic differentiation. | Used to delineate the early vs. late roles of Sox9 [19]. |
| Sox9 siRNA & Lentiviral Overexpression Constructs | For precise loss-of-function (knockdown) and gain-of-function studies to interrogate Sox9's necessity and sufficiency. | Custom siRNA and pLVX-EIF1α-mSox9-IRES-puro lentivirus used in [19]. |
| Chondrogenic Differentiation Media Supplements | Defined components (Insulin, Transferrin, Selenium - ITS) to induce and maintain chondrogenic differentiation in progenitor cells. | 10 µg/ml insulin, 10 µg/ml transferrin, 30 nM sodium selenite used for ATDC5 differentiation [19]. |
| Acellular Cartilage Sheets (ACS) | Scaffolds derived from native cartilage that retain tissue-specific structure and components, providing a biomimetic environment for seeded cells. | Ear (EACS) and Articular (AACS) sheets used to test niche-mimetic properties [21]. |
| Bone Marrow Stromal Cells (BMSCs) | A primary multipotent mesenchymal cell source with high clinical relevance for cartilage regeneration studies. | GFP-labeled porcine BMSCs used for in vivo fate tracking [21]. |
| Assays for Translational Capacity | Functional assays to measure global protein synthesis and ribosome activity, beyond transcriptomics. | SuNSET assay and Polysome profiling [19]. |
Within the broader context of stem cell differentiation research, understanding the specific molecular pathways governing adipogenic lineage commitment is fundamental for advancing regenerative medicine and metabolic disease therapeutics. The process of adipogenesis, through which multipotent mesenchymal stromal cells (MSCs) differentiate into mature adipocytes, is primarily orchestrated by a core transcriptional cascade. This cascade is dominated by the peroxisome proliferator-activated receptor gamma (PPAR-γ) and the CCAAT/enhancer-binding protein (C/EBP) family of transcription factors. These factors do not operate in isolation; they engage in a robust cross-regulatory network that amplifies and stabilizes the differentiation program. Furthermore, this transcriptional core is increasingly understood to be under precise epigenetic control, adding another layer of regulatory complexity. This application note details the molecular mechanisms of this transcriptional control and provides standardized protocols for investigating adipogenic differentiation in vitro, providing researchers with the tools to explore fat cell development in health and disease.
Core Transcriptional Mechanisms
The Master Regulatory Loop: PPAR-γ and C/EBPα
The feed-forward loop between PPAR-γ and C/EBPα is a critical circuit for lineage commitment during adipocytic differentiation. This reciprocal relationship ensures the initiation and maintenance of the adipogenic gene expression program.
- Positive Cross-Regulation: PPAR-γ and C/EBPα positively regulate each other's expression. This mutual reinforcement locks the cell into the adipogenic pathway and maintains the differentiated state [22] [23].
- PPAR-γ as the Proximal Effector: While both factors are essential, PPAR-γ is the indispensable, non-redundant master regulator. Critical loss-of-function experiments demonstrate that C/EBPα has no ability to promote adipogenesis in PPAR-γ-deficient fibroblasts. In contrast, PPAR-γ can induce adipogenesis in C/EBPα-deficient cells, establishing a hierarchical relationship where C/EBPα functions largely through its ability to induce and maintain PPAR-γ expression [24] [25].
- Genome-Wide Cooperativity: On a genomic scale, PPAR-γ and C/EBP factors cooperate extensively. Chromatin immunoprecipitation studies have revealed that in adipocytes, PPAR-γ binds to thousands of genomic sites, and the vast majority of these locations also contain binding motifs for C/EBP factors. Physical colocalization of C/EBPα is observed at a majority of these PPAR-γ-binding regions, and most adipocyte-specific genes are coregulated by both factors, indicating a cooperative orchestration of the adipocyte transcriptome [23].
The following diagram illustrates the core transcriptional network and its regulatory interactions.
Figure 1: The Core Transcriptional Network and its Epigenetic Regulation in Adipogenesis. Early factors C/EBPβ/δ initiate PPAR-γ and C/EBPα expression, which then engage in a positive feed-forward loop to drive the adipogenic program. The methyltransferase PRMT6 represses this loop in precursors via H3R2me2a.
Epigenetic Control of the Transcriptional Circuit
The PPAR-γ–C/EBPα feed-forward loop is repressed in progenitor cells by epigenetic mechanisms, ensuring differentiation only proceeds upon appropriate stimulation.
- PRMT6 as a Key Corepressor: Protein arginine methyltransferase 6 (PRMT6) associates with PPAR-γ on the promoters of both Ppar-γ and C/ebpα in pre-adipocytes, contributing to the repression of their expression. This repressive function is mediated, in part, through PRMT6's ability to deposit the repressive histone mark H3R2me2a, which counteracts activating epigenetic modifications [26].
- Release from Repression upon Differentiation: During the induction of adipocyte differentiation, Prmt6 expression is reduced, and the methyltransferase dissociates from the target promoters. This release of repression allows for the upregulation of PPAR-γ and C/EBPα, thereby establishing the adipocytic gene expression program. Pharmacological inhibition of PRMT6 enhances adipogenesis, highlighting its role as a molecular brake on differentiation [26].
Experimental Protocols for Adipogenic Differentiation
This section provides a detailed methodology for inducing and analyzing adipogenic differentiation in vitro using mesenchymal stromal cells.
Standard Adipogenic Differentiation Protocol
The following protocol is adapted from established methods for inducing adipogenesis in MSC cultures like ST2 or 3T3-L1 cell lines [26].
Materials:
- Cells: Mesenchymal Stromal Cells (e.g., ST2 cells, 3T3-L1 pre-adipocytes, or primary MSCs).
- Basal Medium: Dulbecco's Modified Eagle Medium (DMEM) GlutaMAX, supplemented with 10% Fetal Calf Serum (FCS) and 1% Penicillin/Streptomycin.
- Adipogenic Induction Cocktail:
- 250 nM Dexamethasone
- 450 µM IBMX (3-Isobutyl-1-methylxanthine)
- 1 µM Rosiglitazone (a PPAR-γ agonist)
- 5 µg/ml Insulin
- Maintenance Medium: DMEM with 10% FCS and 5 µg/ml Insulin.
- Reagents for Analysis: Oil-Red-O working solution, 4% Formaldehyde, RNA extraction kit, antibodies for immunoblotting.
Procedure:
- Cell Seeding: Seed cells at a density of 15,000 cells per cm² in standard growth medium and allow them to reach 100% confluency.
- Induction Phase: Replace the growth medium with Adipogenic Induction Medium (basal medium supplemented with the induction cocktail).
- Differentiation Schedule: Culture the cells in induction medium for 3-4 days. This period initiates the transcriptional cascade.
- Maintenance Phase: After the induction phase, switch the culture to Adipogenic Maintenance Medium. Refresh the maintenance medium every 2-3 days.
- Maturation: Continue the culture for a total of 14-21 days to allow for full maturation, including significant lipid accumulation.
Key Considerations:
- The protocol can be adapted for Wharton's Jelly MSCs (WJ-MSCs), though their intrinsic adipogenic potential is lower than that of adipose tissue-derived MSCs (AT-MSCs) [27].
- Optimization may be required for different cell types. For WJ-MSCs, supplementation with 100 µM oleic acid during induction can significantly enhance lipid droplet formation and bring their adipogenic capacity closer to that of AT-MSCs [27].
Protocol for Genetic and Pharmacological Manipulation
To probe the specific roles of transcriptional regulators, genetic and pharmacological tools can be integrated into the differentiation protocol.
- PRMT6 Inhibition: To assess the effect of releasing epigenetic repression, add the selective PRMT6 inhibitor SGC6870 (e.g., at 5 µM concentration) to the induction and/or maintenance medium. A control compound (SGC6870N) should be used in parallel [26].
- Ectopic Gene Expression: To test the sufficiency of a transcription factor, cells can be transduced with retroviruses or lentiviruses expressing the gene of interest (e.g., PPAR-γ or C/EBPα) prior to the induction of differentiation. For example, ectopic expression of PPAR-γ can rescue adipogenesis in PPAR-γ-deficient cells [24] [25].
- Gene Knockdown/Knockout: To test necessity, cells can be transduced with lentiviral vectors expressing specific gRNAs (e.g., in a lentiCRISPRv2 backbone) to knock out genes of interest, followed by selection and differentiation [26].
Analysis of Differentiation Efficiency
Rigorous assessment of adipogenic differentiation is crucial. The table below summarizes the key methods and their applications.
Table 1: Standard Methods for Assessing Adipogenic Differentiation
| Method | Target / Principle | Procedure Summary | Key Output |
|---|---|---|---|
| Oil-Red-O Staining | Staining of neutral lipid droplets in fixed cells. | Fix cells (4% formaldehyde), stain with Oil-Red-O solution, wash, and image. For quantification, elute dye with isopropanol and measure absorbance at 500 nm [26]. | Qualitative visualization and quantitative measurement of lipid accumulation. |
| Triglyceride (TG) Content Assay | Quantitative measurement of intracellular TG. | Use commercial kits based on enzymatic reactions to solubilize and measure TG content, normalized to total cellular protein [27]. | Quantitative, normalized data on lipid storage. |
| Gene Expression Analysis (RT-qPCR) | mRNA levels of adipogenic markers. | Extract RNA, synthesize cDNA, perform qPCR with gene-specific primers. Normalize to housekeeping genes (e.g., TBP) and analyze via ΔΔCT method [26]. | Expression dynamics of key transcriptional regulators and adipocyte genes. |
| Protein Analysis (Immunoblotting) | Protein levels of key transcription factors. | Prepare whole-cell extracts, separate proteins by SDS-PAGE, transfer to membrane, and probe with specific antibodies (e.g., for PPAR-γ, C/EBPα) [26] [24]. | Confirmation of protein expression and post-translational modifications. |
| Chromatin Immunoprecipitation (ChIP) | Transcription factor binding and histone modifications at genomic loci. | Cross-link proteins to DNA, shear chromatin, immunoprecipitate with specific antibody (e.g., anti-PPAR-γ, anti-H3R2me2a), reverse cross-links, and purify DNA for qPCR or sequencing [26] [23]. | Direct evidence of in vivo transcription factor occupancy and epigenetic states. |
Table 2: Key Research Reagent Solutions for Adipogenesis Studies
| Reagent / Resource | Function in Adipogenesis Research | Example & Notes |
|---|---|---|
| PPAR-γ Agonists | Potent chemical inducers of differentiation; activate the master regulator. | Rosiglitazone and Troglitazone are commonly used. Rosiglitazone is included in standard induction cocktails [26] [24]. |
| Small Molecule Inducers | Activate early signaling pathways that initiate the transcriptional cascade. | Dexamethasone (glucocorticoid receptor agonist), IBMX (phosphodiesterase inhibitor that elevates cAMP) [26] [27]. |
| Key Antibodies | Detection of proteins via immunoblotting, immunofluorescence, or ChIP. | Anti-PPAR-γ, Anti-C/EBPα, Anti-FABP4/aP2 (mature adipocyte marker), Anti-PRMT6, Anti-H3R2me2a [26] [23]. |
| Genetic Tools | For gain-of-function and loss-of-function studies. | Lentiviral/Retroviral Vectors for overexpression (e.g., of PPAR-γ) [24] [25] or CRISPR-Cas9 systems for gene knockout (e.g., using lentiCRISPRv2) [26]. |
| Epigenetic Inhibitors | To probe the role of specific epigenetic modifiers. | SGC6870: A selective, small-molecule inhibitor of PRMT6 [26]. |
Advanced Applications and Protocol Optimization
Understanding the core pathway enables researchers to optimize protocols for specific applications and cell types.
- Optimizing WJ-MSC Differentiation: The intrinsic lipid profile of MSCs can influence their differentiation potential. Lipidomic analysis reveals that WJ-MSCs have a different triglyceride profile compared to AT-MSCs. Supplementing the standard adipogenic induction medium with 100 µM oleic acid (OA), a mono-unsaturated fatty acid, significantly enhances lipid droplet formation and upregulates adipogenic markers in WJ-MSCs, making their differentiation efficiency more comparable to AT-MSCs [27].
- Transcriptome Analysis: For a systems-level understanding, RNA sequencing (RNA-seq) can be employed to analyze the global gene expression profile during differentiation. This approach has been instrumental in identifying key molecules, signaling pathways, and biological processes at different time points of adipogenesis [28].
- Lineage Commitment Studies: The commitment of MSCs to adipogenic or osteogenic lineages is a tightly balanced process. Inhibition of the ERK signaling pathway, for instance, can block osteogenic differentiation and simultaneously promote adipogenic differentiation, indicating that molecular switches can redirect cell fate [29].
The following workflow diagram integrates both standard and advanced approaches to studying adipogenesis.
Figure 2: Experimental Workflow for Adipogenesis Research. The core differentiation protocol can be complemented with genetic, epigenetic, and media optimization strategies to address specific research questions.
The transcriptional control of adipogenesis via PPAR-γ and C/EBPs represents a paradigm of cell lineage specification. The precise interplay between these transcription factors, fine-tuned by epigenetic regulators like PRMT6, ensures proper fat cell development. The protocols and tools detailed in this application note provide a robust foundation for researchers to investigate this process, from foundational mechanistic studies to the development of novel therapeutic strategies for metabolic disease and the advancement of soft tissue engineering in regenerative medicine.
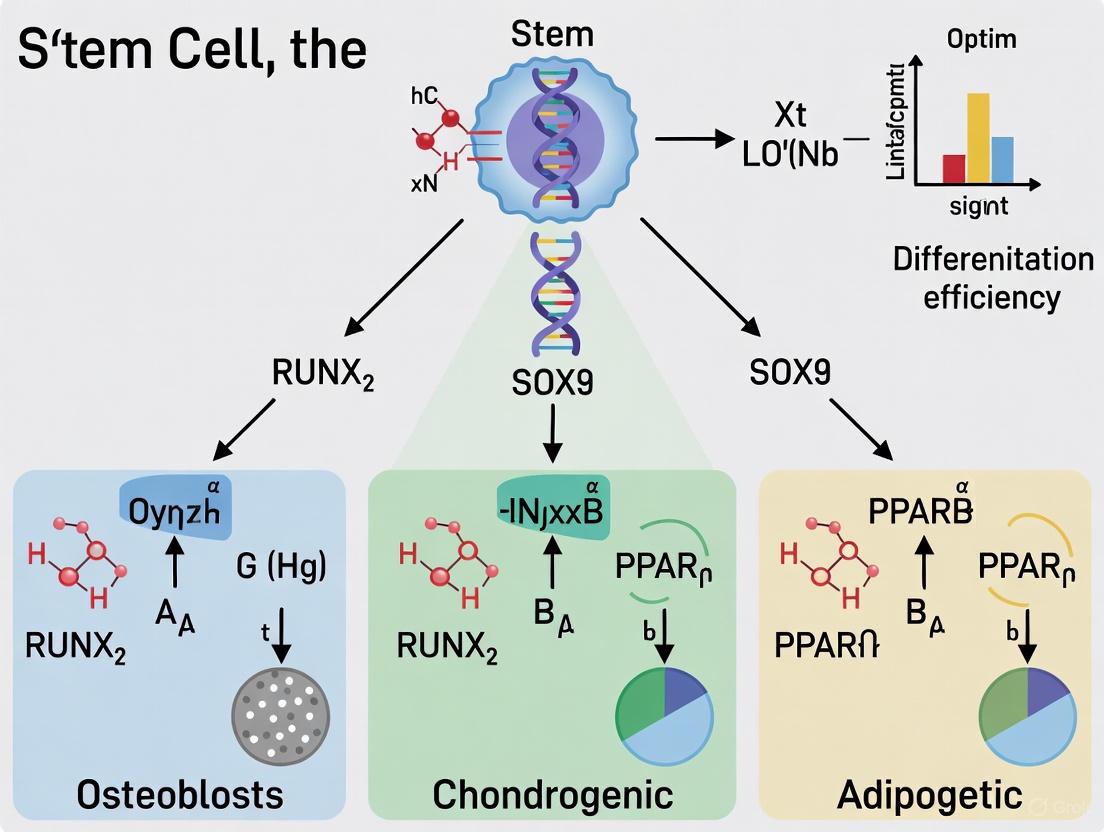
The lineage commitment of mesenchymal stem cells (MSCs) to either osteogenic or adipogenic fates represents a critically balanced process in skeletal homeostasis and whole-body metabolism [30]. As common progenitor cells, MSCs undergo delicately regulated differentiation programs where activation of one lineage often occurs at the expense of the other [31] [30]. This reciprocal relationship is maintained through an intricate network of transcription factors, signaling pathways, and epigenetic modifications that collectively determine cellular fate decisions.
Understanding this balance has significant pathophysiological implications. Aging, obesity, and osteoporosis are frequently characterized by a shift in this equilibrium, with increased bone marrow adiposity coinciding with decreased bone formation [30] [32]. The molecular machinery governing this switch involves core transcription factors including PPARγ2 for adipogenesis and Runx2 for osteogenesis, which often exhibit mutually antagonistic relationships [31]. Additionally, key signaling pathways such as Wnt/β-catenin, BMP, and TGF-β play pivotal roles in directing MSC fate commitment [33] [34].
This application note provides a comprehensive overview of the molecular regulation of osteogenic and adipogenic differentiation, detailed experimental protocols for studying these processes, and key methodological considerations for researchers investigating MSC lineage specification.
Molecular Mechanisms Governing Lineage Specification
Core Transcription Factors and Their Cross-Regulation
The differentiation of MSCs into osteoblasts and adipocytes is governed by two master transcription factors that demonstrate a mutually antagonistic relationship.
Table 1: Core Transcription Factors in Osteogenesis and Adipogenesis
| Transcription Factor | Primary Lineage | Key Target Genes | Antagonistic Mechanisms |
|---|---|---|---|
| Runx2 | Osteogenesis | Osteocalcin (OC), Bone Sialoprotein (BSP), Alkaline Phosphatase (APL) | Suppresses PPARγ2 transactivation; induces osteogenic genes |
| PPARγ2 | Adipogenesis | Fatty Acid-Binding Protein 4 (FABP4), Lipoprotein Lipase (LPL) | Inhibits Runx2-mediated transcription of osteocalcin |
| sLZIP | Regulatory | HDAC3, PPARγ2 complex | Interacts with PPARγ2 and HDAC3 to suppress PPARγ2 activity while enhancing Runx2 |
The PPARγ2-Runx2 axis forms the core regulatory circuit governing the adipogenesis-osteogenesis balance [31]. PPARγ2 activation not only promotes adipogenic differentiation but also directly inhibits osteoblast differentiation by suppressing Runx2 transcriptional activity [31]. Conversely, Runx2 expression inhibits adipogenesis by interfering with PPARγ2 function. This reciprocal inhibition ensures that MSCs commit predominantly to one lineage.
Recent research has identified regulatory proteins that modulate this core circuit. The small leucine zipper protein (sLZIP) acts as a novel PPARγ2 modulator by interacting with PPARγ2 and recruiting histone deacetylase 3 (HDAC3) to form a corepressor complex [31]. This complex suppresses PPARγ2 transcriptional activity, thereby inhibiting adipogenesis while simultaneously promoting osteogenesis through enhanced Runx2 activity [31]. Transgenic mice overexpressing sLZIP demonstrate enhanced bone mass and density, confirming its role in directing MSC fate toward osteogenesis.
Signaling Pathways in Lineage Determination
Multiple evolutionarily conserved signaling pathways interact to fine-tune the balance between osteogenic and adipogenic differentiation.
Table 2: Signaling Pathways in MSC Lineage Specification
| Signaling Pathway | Effect on Osteogenesis | Effect on Adipogenesis | Key Molecular Mediators |
|---|---|---|---|
| Wnt/β-catenin | Promotes | Inhibits | LRP5/6, β-catenin, GSK3β, TAZ |
| TGF-β/BMP | Context-dependent promotion | Context-dependent inhibition | Smads, MAPK, Runx2, PPARγ |
| Hedgehog | Promotes | Inhibits | Gli proteins, Smo, Ptch |
| Notch | Complex (inhibitory or promotional) | Inhibits | Hes, Hey, PPARγ |
Wnt/β-catenin Signaling
The canonical Wnt/β-catenin pathway serves as a potent promoter of osteogenesis while simultaneously inhibiting adipogenesis [30]. Wnt ligands binding to Frizzled receptors and LRP5/6 co-receptors stabilize β-catenin, which translocates to the nucleus and activates osteogenic target genes including Runx2 [34]. Additionally, Wnt signaling activates the transcriptional coactivator TAZ, which enhances Runx2-dependent gene transcription while suppressing PPARγ-mediated adipogenic differentiation [30]. Recent research has identified Mapk7 as a novel activator of Wnt signaling, which enhances osteogenesis and suppresses adipogenesis by phosphorylating Lrp6 at Ser1490, thereby stabilizing β-catenin [32].
TGF-β/BMP Signaling
The TGF-β/BMP pathway exhibits complex, context-dependent effects on MSC differentiation [33]. BMP2 demonstrates concentration-dependent effects: at low doses (50 ng/mL) it can promote adipogenesis, while at higher doses (500 ng/mL) it strongly promotes osteogenic differentiation [30] [33]. TGF-β1 and TGF-β3 generally inhibit adipogenic differentiation while promoting chondrogenesis [33]. The adipogenesis inhibition occurs primarily through Smad3, which associates with C/EBPβ and C/EBPδ to suppress PPARγ expression [33].
Figure 1: Molecular regulation of the adipogenesis-osteogenesis balance. Key transcription factors PPARγ and Runx2 demonstrate mutual inhibition, while signaling pathways exert directional control on lineage commitment.
Epigenetic Regulation and Omics Characteristics
Epigenetic modifications play a pivotal role in mediating heritable changes in gene expression without altering the DNA sequence during MSC differentiation [35]. Advances in omics technologies have enhanced our understanding of ADSC molecular profiles through transcriptomic, proteomic, and lipidomic analyses [35].
Transcriptomic studies using single-cell RNA sequencing have revealed considerable heterogeneity within ADSC populations [35]. Distinct subpopulations exhibit different lineage commitment capabilities, with one subcluster expressing high levels of adipogenic markers (Pparg, Cd36) representing committed preadipocytes, while another fraction characterized by Cd142 and Abcg1 expression negatively regulates adipogenesis through paracrine mechanisms [35].
Proteomic analyses have identified distinct protein expression patterns between ADSCs and BMSCs. ADSCs exhibit proteins associated with biological oxidation, nucleobase biosynthesis, and vitamin metabolism, suggesting higher basal metabolic activity, while BMSCs show elevated expression of proteins involved in extracellular matrix organization and cell-matrix interactions [35].
Lipidomics studies have revealed that ADSCs possess a distinctive and more diverse phospholipid profile compared to other cell types, with specific species such as phosphatidylglycerol (PG) 40:7 and phosphatidylethanolamine (PE) O-36:3 detected exclusively in ADSCs [35]. Sphingomyelins (SMs) are also predominantly present in ADSCs, suggesting potential roles for phospholipids and sphingolipids in regulating ADSC biology [35].
Experimental Models and Methodological Approaches
In Vitro Differentiation Protocols
Standardized protocols for inducing and assessing osteogenic and adipogenic differentiation are essential for studying MSC fate decisions.
Osteogenic Differentiation Protocol
Materials:
- C3H10T1/2 MSC line or primary MSCs
- Osteogenic Induction Medium (OIM): DMEM supplemented with 10% FBS, 10 mM β-glycerophosphate, 50 μM ascorbate-2-phosphate, and 100 nM dexamethasone
- 4% paraformaldehyde (PFA) for fixation
- Alkaline Phosphatase (ALP) staining solution
- Alizarin Red S (ARS) staining solution (2%, pH 4.2)
Procedure:
- Seed MSCs at a density of 10,000 cells/cm² in growth medium and culture until 80% confluent.
- Replace growth medium with Osteogenic Induction Medium (OIM).
- Change the induction medium every 3-4 days for up to 21 days.
- For alkaline phosphatase (ALP) staining, after 7 days of induction, rinse cells with PBS, fix with 4% PFA for 10 minutes, and incubate with ALP staining solution until desired color development [32].
- For mineralization assessment (Alizarin Red S staining), after 21 days of induction, fix cells with 4% PFA for 15 minutes, then stain with 2% ARS solution (pH 4.2) for 20 minutes [32].
- For quantitative analysis, dissolve calcium nodules in DMSO and measure absorbance at 405 nm [32].
Adipogenic Differentiation Protocol
Materials:
- C3H10T1/2 MSC line or primary MSCs
- Adipogenic Induction Medium (AIM): DMEM supplemented with 10% FBS, 0.5 mM 3-isobutyl-1-methylxanthine (IBMX), 1 μM dexamethasone, 10 μM insulin, and 200 μM indomethacin
- Oil Red O (ORO) staining solution (0.5% in isopropanol)
- 4% paraformaldehyde (PFA) for fixation
Procedure:
- Seed MSCs at a density of 20,000 cells/cm² in growth medium until 100% confluent.
- Maintain at confluence for 2-3 days (post-confluence stage).
- Replace growth medium with Adipogenic Induction Medium (AIM).
- After 3-5 days of induction, change to adipogenic maintenance medium (DMEM with 10% FBS and 10 μM insulin) for 2-3 days.
- Repeat induction/maintenance cycles 2-3 times over 10-14 days.
- For lipid droplet visualization, fix cells with 4% PFA for 15 minutes and stain with filtered Oil Red O working solution for 30-60 minutes [36].
- For quantitative analysis, extract stained lipid droplets with 100% isopropanol and measure absorbance at 510 nm [32].
Modulation of Signaling Pathways
Experimental manipulation of key signaling pathways allows researchers to direct MSC fate decisions.
Wnt/β-catenin pathway activation:
- Add 10 nM CHIR99021 (GSK3β inhibitor) to differentiation medium [32]
- Co-culture with L Wnt-3A cells using trans-well systems [32]
- Use recombinant Wnt3a protein (50-100 ng/mL)
TGF-β/BMP pathway modulation:
- For osteogenesis: BMP2 at 500 ng/mL [33]
- For adipogenesis: BMP2 at 50 ng/mL with PPARγ activator [33]
- TGF-β1 at 10 ng/mL for chondrogenesis or adipogenesis inhibition [33]
Mapk7 manipulation:
- Knockout models: Prx1-Cre; Mapk7flox/flox mice [32]
- Overexpression studies using viral vectors
Analysis Methods for Differentiation Assessment
Table 3: Analytical Methods for Assessing MSC Differentiation
| Analysis Type | Method | Key Markers/Targets | Application |
|---|---|---|---|
| Gene Expression | RT-qPCR | Osteogenesis: Runx2, ALP, OCN, Osterix Adipogenesis: PPARγ, C/EBPα, FABP4, adiponectin | Quantitative assessment of lineage-specific gene expression |
| Protein Analysis | Western Blot, Immunofluorescence | Osteogenesis: Runx2, Osterix, Osteocalcin Adipogenesis: PPARγ, FABP4, ACC | Protein level confirmation of differentiation |
| Histochemical Staining | ALP, ARS, ORO | ALP activity (early osteogenesis), Calcium deposition (late osteogenesis), Lipid accumulation (adipogenesis) | Qualitative and semi-quantitative assessment of differentiation extent |
| Flow Cytometry | Surface marker analysis | CD73, CD90, CD105 (positive); CD34, CD45 (negative) | MSC phenotype verification before differentiation |
The Scientist's Toolkit: Research Reagent Solutions
Table 4: Essential Research Reagents for Adipogenesis and Osteogenesis Studies
| Reagent/Category | Specific Examples | Function/Application | Experimental Notes |
|---|---|---|---|
| Cell Lines | C3H10T1/2, 3T3-L1, Primary MSCs | In vitro differentiation models | Primary MSCs require phenotype verification via flow cytometry |
| Induction Cocktails | Dexamethasone, IBMX, Insulin, Indomethacin, β-glycerophosphate, Ascorbate-2-phosphate | Direct lineage-specific differentiation | Cyclic induction recommended for adipogenesis (3-5 day cycles) |
| Signaling Modulators | CHIR99021 (Wnt activator), Recombinant BMP2/TGF-β, SOST antibodies | Pathway-specific manipulation | Concentration-dependent effects observed with BMP2 |
| Staining Reagents | Alizarin Red S, Oil Red O, Alkaline Phosphatase staining kits | Differentiation endpoint assessment | Quantitative extraction protocols available for mineralization and lipid content |
| Antibodies | Anti-Runx2, Anti-PPARγ, Anti-Osteocalcin, Anti-FABP4 | Protein-level confirmation of differentiation | Essential for Western blot and immunofluorescence validation |
Applications and Research Implications
Disease Modeling and Pathophysiological Insights
The inverse relationship between osteogenesis and adipogenesis has significant implications for understanding and treating metabolic bone diseases. In osteoporosis, increased bone marrow adiposity coincides with decreased bone formation, representing a shift in MSC lineage commitment [30] [32]. Similarly, in obesity and type 2 diabetes, dysfunction in ADSC differentiation potential contributes to impaired adipose tissue plasticity and metabolic complications [37].
Notably, the unique regenerative capacity of Acomys cahirinus (spiny mice) provides intriguing insights into MSC biology. ADSCs from Acomys cahirinus exhibit enhanced osteogenesis and suppressed adipogenesis compared to Mus musculus, which is linked to their exceptional tissue regeneration capabilities but potentially limits their adipose tissue self-renewal [37]. This model system offers opportunities to identify novel regulatory mechanisms that could be therapeutically targeted.
Therapeutic Targeting and Tissue Engineering
Understanding the molecular basis of the adipogenesis-osteogenesis balance enables development of targeted therapeutic strategies. Several approaches show promise:
Wnt pathway modulation:
- Anti-SOST antibodies (Romosozumab, Blosozumab) to enhance bone formation [34]
- Small molecule GSK3β inhibitors to stabilize β-catenin [32]
Transcription factor modulation:
- PPARγ partial agonists to minimize adverse effects on bone [31]
- sLZIP-based approaches to simultaneously inhibit adipogenesis and promote osteogenesis [31]
Biomaterial applications:
- Surface topography engineering with specific ridge patterns (2 μm ridges enhance osteogenesis, 15 μm ridges increase adipogenesis) [38]
- Nanostructured substrates (650 nm periodicity) that enhance both osteogenic and adipogenic differentiation when combined with induction media [38]
Figure 2: Experimental workflow for studying adipogenesis-osteogenesis balance. A comprehensive approach combining differentiation assays with pathway modulation and multi-modal analysis enables mechanistic insights.
Concluding Remarks
The reciprocal relationship between osteogenesis and adipogenesis represents a fundamental aspect of MSC biology with far-reaching implications for regenerative medicine, metabolic disease treatment, and tissue engineering. The core regulatory circuit centered on PPARγ2 and Runx2 antagonism, modulated by various signaling pathways and epigenetic mechanisms, provides a sophisticated control system for MSC fate decisions.
Methodologically, robust protocols for inducing and assessing differentiation, combined with targeted pathway modulation approaches, enable detailed investigation of this balance. The continued development of specific reagents and advanced biomaterials that can direct MSC lineage commitment holds promise for novel therapeutic strategies for conditions characterized by disruption of the adipogenesis-osteogenesis equilibrium, such as osteoporosis, obesity, and diabetes.
Future research directions should focus on understanding the temporal dynamics of lineage commitment decisions, the role of epigenetic memory in MSC differentiation, and the development of spatiotemporal control systems for precise regulation of MSC fate in therapeutic contexts.
Application Notes
Stem cell differentiation into osteogenic, chondrogenic, and adipogenic lineages is a tightly regulated process guided by dynamic changes in the transcriptome. The emergence of high-throughput RNA sequencing (RNA-seq) technologies, particularly single-cell RNA-seq (scRNA-seq), has revolutionized our ability to decode these complex molecular events. Long non-coding RNAs (lncRNAs), once considered "genomic junk," are now recognized as vital regulators of gene expression during cell fate determination. This article explores how transcriptomic insights, especially those concerning non-coding RNAs, are shaping our understanding of lineage commitment in stem cell biology, with significant implications for regenerative medicine and therapeutic development.
The Transcriptomic Landscape of Stem Cell Differentiation
Transcriptome analysis during stem cell differentiation reveals precisely timed molecular programs that guide lineage specification. Mesenchymal stem/stromal cells (MSCs) possess the capacity to differentiate into adipogenic, osteogenic, and chondrogenic lineages when stimulated under appropriate conditions, making them a primary model for studying lineage commitment [28]. The transcriptome of a stem cell represents the complete set of RNA molecules that dictate its functional state, with lineage commitment directed by specific gene expression profiles and their complex interactions [28].
Single-cell transcriptomic analyses have been particularly transformative, revealing that exit from pluripotency marks the start of a lineage transition accompanied by a transient phase of increased susceptibility to lineage-specifying signals [39]. During retinoic acid-driven differentiation of mouse embryonic stem cells (mESCs), researchers observed a sharp increase in gene expression variability between 24-48 hours of exposure, coinciding with the exit from pluripotency and the beginning of lineage commitment [39]. This period of increased transcriptional heterogeneity may represent a critical window where cell fate decisions are most malleable to external cues.
Non-Coding RNAs as Master Regulators of Cell Fate
Long non-coding RNAs (lncRNAs), defined as RNA transcripts exceeding 200 nucleotides without protein-coding capacity, have emerged as crucial regulators of stem cell pluripotency and differentiation [40] [41]. These molecules exert their regulatory effects through diverse mechanisms depending on their subcellular localization and interacting partners.
In the cytoplasm, lncRNAs typically regulate mRNA stability, mediate translation, and function as competing endogenous RNAs. In contrast, nuclear lncRNAs more commonly influence chromatin architecture and transcriptional activity through interactions with DNA, RNA, and proteins [40]. For example, XIST lncRNA recruits polycomb repressive complexes to trigger histone modifications that silence gene transcription, while MALAT1 functions as a scaffold molecule in nuclear speckles to regulate splicing [40].
During embryonic stem cell-derived cardiomyocyte differentiation, lncRNAs demonstrate highly dynamic expression patterns, with the largest group enriched specifically in ESCs [40]. Systematic analysis of lncRNA expression across four critical developmental stages revealed that differentially expressed lncRNAs group into six distinct clusters, suggesting specialized functions at different differentiation timepoints [40].
Epitranscriptomic Modifications Fine-Tune LncRNA Function
The regulatory capacity of lncRNAs is further refined by post-transcriptional modifications, particularly N6-methyladenosine (m6A) - the most abundant RNA modification identified in mRNA [40]. This modification can significantly influence lncRNA functionality by recruiting specific "reader" proteins.
Research has demonstrated that m6A residues on lncRNAs recruit nuclear reader proteins like YTH domain containing 1 (YTHDC1), which is required for XIST-mediated transcriptional silencing [40]. During ESC differentiation, a subset of lncRNAs shows significant m6A modification and direct interaction with YTHDC1 [40]. Notably, the ESC-specific lncRNA Gm2379 is dramatically upregulated in response to m6A or Ythdc1 depletion, and its own depletion dysregulates pluripotency genes and those required for germ layer formation [40]. This epitranscriptomic regulation represents an additional layer of control in stem cell fate decisions.
Signaling Pathways in Lineage Commitment
The integration of external and internal signals guides fate commitment in differentiating pluripotent cells [39]. The MAPK signaling pathway, particularly ERK activation, plays a pivotal role in determining whether human MSCs commit to osteogenic or adipogenic lineages [29].
During osteogenic differentiation, treatment with osteogenic supplements induces a sustained phase of ERK activation from day 7 to day 11 that coincides with differentiation, before decreasing to basal levels [29]. JNK activation occurs later (day 13-17) and associates with extracellular matrix synthesis and calcium deposition - hallmark processes of bone formation [29]. Significantly, inhibition of ERK activation blocks osteogenic differentiation in a dose-dependent manner and redirects fate toward adipogenic differentiation [29]. This demonstrates how the same signaling pathway can act as a molecular switch between alternative lineage commitments.
Table 1: Key Signaling Pathways in Mesenchymal Stem Cell Lineage Commitment
| Pathway | Role in Osteogenesis | Role in Adipogenesis | Key Regulators |
|---|---|---|---|
| MAPK/ERK | Sustained activation promotes differentiation [29] | Inhibition redirects from osteogenesis [29] | ERK, JNK, p38 |
| Retinoic Acid | Induces neuroectodermal and XEN lineages [39] | Suppresses mesodermal genes [39] | RA receptors |
| m6A Modification | Regulates lncRNAs guiding lineage commitment [40] | Potential role through lncRNA regulation [40] | METTL3, YTHDC1 |
Technological Advances in Transcriptome Analysis
The evolution from hybridization-based microarrays to next-generation RNA sequencing has dramatically enhanced our ability to study stem cell differentiation [28]. RNA-seq provides precise measurements of transcript abundance with single-base resolution, can distinguish splicing isoforms, and does not require prior knowledge of genes present in the analyzed genome [28]. This technological advancement has been particularly valuable for identifying novel non-coding RNA species, including various classes of regulatory lncRNAs [28].
Single-cell RNA sequencing has further revolutionized the field by enabling researchers to characterize heterogeneity within stem cell populations and trace transcriptional dynamics during lineage commitment [39] [42]. This approach has revealed that cells in the neural stem cell lineage exist on a continuum through activation and differentiation processes, with rare intermediate states possessing distinct molecular profiles [42]. Pseudotemporal ordering of scRNA-seq data can reconstruct developmental trajectories and identify putative regulators of cell fate decisions [42].
More recently, high-resolution spatial transcriptomics has enabled molecular identification of cell types based on spatially resolved gene expression profiles in developing tissues [43]. Integrating scRNA-seq with spatial transcriptomics during craniofacial development has revealed that mesenchymal lineage establishment occurs prior to anatomical structure formation, with heterogeneous progenitor populations already activating early lineage-specific markers at the onset of development [43].
Table 2: Transcriptomic Technologies for Studying Lineage Commitment
| Technology | Key Applications | Advantages | Limitations |
|---|---|---|---|
| Bulk RNA-seq | Population-level expression profiling [28] | Detects overall expression patterns; cost-effective [28] | Masks cellular heterogeneity [39] |
| Single-cell RNA-seq | Resolving cellular heterogeneity; trajectory inference [39] [42] | Reveals rare cell states; reconstructs differentiation paths [42] | Higher cost; technical noise [39] |
| Spatial Transcriptomics | Mapping gene expression to tissue location [43] | Preserves spatial context; links location to fate [43] | Lower resolution than scRNA-seq [43] |
Experimental Protocols
Protocol 1: In Vitro Differentiation of Human MSCs into Osteogenic, Chondrogenic, and Adipogenic Lineages
This protocol describes standard methods for differentiating bone marrow-derived MSCs (BM-MSCs) into three key mesodermal lineages, based on established characterization criteria that include plastic-adherence capacity, defined epitope profile, and multipotent differentiation capability [44].
Materials
- Human BM-MSCs: Isolated from bone marrow aspirates
- Basal medium: α-MEM supplemented with 10% fetal bovine serum (FBS), 100 U/mL penicillin, and 100 μg/mL streptomycin
- Osteogenic supplements: 10 mM β-glycerophosphate, 50 μM ascorbic acid, and 100 nM dexamethasone
- Adipogenic supplements: 0.5 mM 3-isobutyl-1-methylxanthine (IBMX), 1 μM dexamethasone, 10 μM insulin, and 200 μM indomethacin
- Chondrogenic supplements: 10 ng/mL TGF-β3, 100 nM dexamethasone, 50 μg/mL ascorbic acid-2-phosphate, 40 μg/mL proline, and 1% ITS+ premix
Osteogenic Differentiation Procedure
- Seed BM-MSCs at a density of 5 × 10^3 cells/cm² in basal medium and culture until 70% confluent
- Replace medium with osteogenic induction medium containing supplements
- Change medium every 3-4 days for 21 days
- Monitor differentiation by alkaline phosphatase staining (after 7-10 days) and Alizarin Red S staining for calcium deposition (after 21 days)
- For molecular analysis, harvest cells at various timepoints for RNA extraction and transcriptomic analysis
Adipogenic Differentiation Procedure
- Seed BM-MSCs at a density of 2 × 10^4 cells/cm² in basal medium
- At 100% confluence, induce differentiation with adipogenic induction medium for 3 days
- Switch to adipogenic maintenance medium (basal medium with 10 μM insulin only) for 1-3 days
- Repeat this cycle 3-5 times
- Confirm adipogenesis by Oil Red O staining of lipid vacuoles after 14-21 days
Chondrogenic Differentiation Procedure
- Harvest BM-MSCs and pellet 2.5 × 10^5 cells by centrifugation at 500 g for 10 minutes
- Maintain cell pellets in chondrogenic induction medium in polypropylene tubes
- Change medium every 3-4 days for 28 days
- Assess chondrogenesis by Alcian Blue staining for sulfated proteoglycans or immunohistochemistry for collagen type II
Protocol 2: Single-Cell RNA-seq Analysis of Differentiation Dynamics
This protocol outlines an approach for capturing transcriptome dynamics during stem cell differentiation at single-cell resolution, based on methodologies successfully applied to study retinoic acid-driven differentiation of mouse ESCs [39] and adult neural stem cells [42].
Materials
- Single-cell RNA barcoding and sequencing (SCRB-seq) reagents [39]
- Cell suspension at appropriate differentiation timepoints
- Next-generation sequencing platform
- Bioinformatics tools for data analysis (Seurat, Monocle, etc.)
Procedure
Sample Collection and Preparation:
- Collect cells at multiple timepoints during differentiation (e.g., 0, 12, 24, 36, 48, 72, 96 hours)
- Prepare single-cell suspension using enzymatic dissociation
- Ensure cell viability >90% before proceeding
Single-Cell Library Preparation:
- Use SCRB-seq method to barcode individual cells [39]
- Perform reverse transcription and cDNA amplification
- Prepare sequencing libraries with appropriate adapters
Sequencing and Data Processing:
- Sequence libraries to a depth of 5-50 thousand reads per cell
- Align reads to reference genome using HISAT2 or similar aligner [41]
- Generate gene expression matrices
Data Analysis:
- Perform quality control to remove low-quality cells and doublets
- Normalize data using log normalization or SCTransform
- Identify highly variable genes for downstream analysis
- Conduct principal component analysis (PCA) to reduce dimensionality
- Cluster cells using graph-based clustering algorithms
- Visualize results using t-distributed stochastic neighbor embedding (t-SNE) or Uniform Manifold Approximation and Projection (UMAP)
Lineage Trajectory Reconstruction:
- Order cells along pseudotime using tools like Monocle
- Identify genes with dynamic expression during differentiation
- Group genes into co-expression modules
Protocol 3: Functional Validation of lncRNAs in Lineage Commitment
This protocol describes methods for investigating the functional role of specific lncRNAs in stem cell differentiation, based on approaches used to characterize lncRNAs like Gm2379 in mESC fate decisions [40].
Materials
- Lentiviral vectors for shRNA-mediated knockdown
- Lipofectamine or similar transfection reagent
- qPCR reagents for expression validation
- Antibodies for immunostaining of lineage markers
Procedure
Identification of Candidate lncRNAs:
- Analyze RNA-seq data to identify lncRNAs differentially expressed during differentiation
- Select candidates based on expression dynamics and potential relevance to studied lineage
Loss-of-Function Studies:
- Design 2-3 shRNAs targeting different regions of the candidate lncRNA
- Clone shRNA sequences into lentiviral vectors (e.g., pRSI9-U6-(sh)-UbiC-TagRFP-2A-Blasticidin) [40]
- Package lentiviral particles using Lenti-X 293T cells
- Transduce stem cells with lentivirus and select with appropriate antibiotics
- Validate knockdown efficiency by qRT-PCR
Phenotypic Characterization:
- Differentiate knockdown and control cells along specific lineages
- Assess differentiation efficiency using lineage-specific stains
- Analyze expression of key lineage markers by qRT-PCR
- For spatial assessment, perform RNA fluorescence in situ hybridization (FISH)
Mechanistic Studies:
- Perform RNA immunoprecipitation (RIP) to identify protein interaction partners
- Conduct chromatin isolation by RNA purification (ChIRP) to map genomic binding sites
- Assess epigenetic changes at potential target genes
Signaling Pathways and Regulatory Networks
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Research Reagents for Studying Transcriptome Dynamics in Lineage Commitment
| Reagent/Category | Specific Examples | Function/Application |
|---|---|---|
| Cell Culture Media | KnockOut Serum Replacement, N2B27 medium [40] [39] | Supports pluripotent stem cell maintenance and differentiation |
| Differentiation Inducers | Retinoic acid, β-glycerophosphate, TGF-β3, IBMX [39] [44] | Directs stem cells toward specific lineages (osteogenic, chondrogenic, adipogenic) |
| Lineage Markers | Alkaline phosphatase (osteogenic), Oil Red O (adipogenic), Alcian Blue (chondrogenic) [44] | Histochemical staining to confirm differentiation efficiency |
| Molecular Biology Tools | shRNAs for lncRNA knockdown, YTHDC1 antibodies, m6A-specific antibodies [40] | Functional validation of non-coding RNAs and epitranscriptomic modifications |
| Sequencing Reagents | SCRB-seq barcoding reagents, reverse transcriptase, sequencing adapters [39] | Single-cell RNA library preparation for transcriptome analysis |
| Bioinformatics Software | Seurat, Monocle, HISAT2, StringTie [41] [43] [42] | Data processing, normalization, clustering, and trajectory analysis |
The integration of transcriptomic technologies, particularly single-cell and spatial RNA sequencing, with functional studies of non-coding RNAs has dramatically advanced our understanding of lineage commitment in stem cell biology. The dynamic interplay between protein-coding genes, lncRNAs, and epitranscriptomic modifications creates a multi-layered regulatory network that guides cell fate decisions. These insights not only enhance our fundamental knowledge of developmental biology but also provide new avenues for therapeutic intervention in regenerative medicine, disease modeling, and drug development. As transcriptomic technologies continue to evolve, particularly in resolution and throughput, we can anticipate even deeper insights into the molecular choreography of stem cell differentiation.
From Bench to Bedside: Innovative Induction Techniques and Tissue Engineering Strategies
Within the field of regenerative medicine, the directed differentiation of human Mesenchymal Stem/Stromal Cells (hMSCs) into specific lineages represents a cornerstone for both basic research and therapeutic development [28]. These primary, multipotent cells are capable of differentiating into osteogenic (bone), chondrogenic (cartilage), and adipogenic (fat) lineages when stimulated under appropriate biochemical conditions [28] [29]. This application note provides detailed, structured protocols for the traditional biochemical induction of these three lineages, framed within the context of a broader thesis on stem cell differentiation. It is designed to equip researchers, scientists, and drug development professionals with the standardized methodologies necessary to ensure rigor, reproducibility, and translational relevance in their work. The fundamental principle guiding these processes is the careful manipulation of the cellular microenvironment through specific signaling molecules and culture conditions to direct cell fate decisions [28]. Understanding the competitive and reciprocal relationship between these pathways, particularly between osteogenesis and adipogenesis, is crucial for developing therapies for conditions like osteoporosis, where an imbalance in these lineages leads to pathology [45].
The Scientist's Toolkit: Essential Reagents for Lineage Differentiation
The following table catalogs the core reagents essential for initiating and maintaining the differentiation of hMSCs. Sourcing high-quality components is critical for experimental success.
Table 1: Key Research Reagent Solutions for hMSC Differentiation
| Reagent/Solution | Primary Function in Differentiation | Example Components |
|---|---|---|
| Basal Growth Medium | Supports hMSC expansion and maintenance prior to induction. | Dulbecco’s Modified Eagle’s Medium (DMEM) or α-MEM, supplemented with Fetal Bovine Serum (FBS), penicillin, and streptomycin [45]. |
| Osteogenic Induction Medium | Drives commitment to the osteoblastic lineage, promoting matrix mineralization. | Dexamethasone, Glycerol 2-phosphate, Ascorbic Acid [45]. |
| Adipogenic Induction Medium | Induces formation of lipid-laden adipocytes. | Insulin, Dexamethasone, 3-Isobutyl-1-methylxantine (IBMX), Indomethacin [45]. |
| Chondrogenic Induction Medium | Promotes the formation of cartilaginous tissue and matrix. | Transforming Growth Factor-Beta (TGF-β), Insulin, Ascorbic Acid, Dexamethasone (often in pellet or micromass culture). |
| Specific Markers for Validation | Enables confirmation of successful differentiation via qRT-PCR, staining, or immunofluorescence. | Osteogenesis: Alkaline Phosphatase (ALP), Runt-related transcription factor 2 (RUNX2). Adipogenesis: Peroxisome Proliferator-Activated Receptor γ2 (PPARγ2), Lipoprotein Lipase (LPL). Chondrogenesis: Collagen type II, Aggrecan (ACAN) [28] [29] [45]. |
Detailed Experimental Protocols
Cell Culture and Seeding
- Cell Source: Isolate hMSCs from bone marrow aspirate or use commercially available primary hMSCs. The International Society for Cellular Therapy (ISCT) defines hMSCs by specific surface marker expression (>95% positive for CD105, CD73, CD90; <2% positive for CD45, CD34, etc.) and functional multipotency [28].
- Expansion Culture: Maintain hMSCs in Basal Growth Medium (e.g., DMEM with 10% FBS and 1% penicillin/streptomycin) at 37°C in a 5% CO₂ atmosphere. Replace the medium every 2-3 days [45].
- Seeding for Differentiation: At passage 2-4, harvest hMSCs using trypsin/EDTA and seed them into multi-well culture plates at a recommended density of 1.5 × 10⁴ cells/cm² [45].
- Initiation of Induction: Allow cells to attach and grow in Basal Growth Medium until they reach 90-100% confluence. At this point, replace the growth medium with the specific induction medium.
Quantitative Composition of Induction Media
The precise formulation of induction media is critical. The table below summarizes the key components and their concentrations for each lineage.
Table 2: Composition of Traditional Biochemical Induction Media
| Lineage | Key Inducing Factors | Typical Concentration | Reported Yield/Outcome Markers |
|---|---|---|---|
| Osteogenic | Dexamethasone [45] | 100 nM [45] | Upregulation of ALP, Osteocalcin; Calcium deposition observed from day 13-17 [29]. |
| Glycerol 2-phosphate [45] | 10 mM [45] | ||
| Ascorbic Acid [45] | 50 μM [45] | ||
| Adipogenic | Insulin [45] | 5 μg/mL [45] | Expression of PPARγ2, aP2, LPL; visible lipid droplet accumulation [29]. |
| Dexamethasone [45] | 1 μM [45] | ||
| 3-Isobutyl-1-methylxantine (IBMX) [45] | 500 μM [45] | ||
| Indomethacin [45] | 50 μM [45] | ||
| Chondrogenic | Transforming Growth Factor-β (TGF-β) | Commonly 10 ng/mL | Upregulation of Collagen type II, Aggrecan; synthesis of sulfated proteoglycan-rich matrix. |
Protocol Maintenance and Validation
- Medium Refreshment: Replace the induction medium every 2-3 days to ensure a consistent supply of fresh inducing factors and nutrients [45].
- Differentiation Timeline: The initial lineage commitment phase typically occurs within the first 2-4 days, with stable lineage-specific markers becoming expressed and mature matrix production occurring over 2-4 weeks [45].
- Validation: Success of differentiation must be confirmed using a combination of techniques:
- Gene Expression: Quantitative RT-PCR for lineage-specific genes (e.g., ALP, PPARγ2, COL2A1) [45].
- Histochemical Staining: Alizarin Red S (mineralization for osteogenesis), Oil Red O (lipid droplets for adipogenesis), Alcian Blue or Safranin O (sulfated proteoglycans for chondrogenesis).
- Immunofluorescence: Staining for specific proteins like Osteocalcin (osteogenesis) or Collagen type II (chondrogenesis).
Signaling Pathways Governing Lineage Commitment
The biochemical inducers in the media exert their effects by activating or inhibiting specific intracellular signaling pathways, which in turn control the transcriptional programs that define cell fate. Key pathways and their interactions are illustrated below.
The diagram above illustrates the key signaling pathways activated by traditional biochemical inducers. A critical regulatory mechanism involves the Mitogen-activated Protein Kinase (MAPK) pathway, specifically ERK activation. Sustained ERK activation from day 7 to 11 is required for osteogenic differentiation. Inhibition of ERK not only blocks osteogenesis but can actually divert hMSCs toward the adipogenic lineage, demonstrating a reciprocal regulatory switch between these two fates [29]. This underscores the importance of precise temporal control in induction protocols.
Transcriptome analyses have further elucidated other critical early-responder pathways. During the initial stages of commitment (days 2-4), the FoxO signaling pathway (involving FoxO3, IL6, CAT) is crucial for osteogenesis, while the Rap1 signaling pathway ( involving VEGFA, FGF2) is more significant for adipogenesis [45]. The PI3K-Akt signaling pathway may serve as a latent mechanism involved in the initiation of differentiation into multiple lineages [45].
Integrated Experimental Workflow
A typical workflow for a differentiation experiment, from cell preparation to final analysis, integrates all the components previously described and is outlined below.
The traditional biochemical induction protocols detailed herein provide a reliable foundation for directing hMSC fate toward osteogenic, adipogenic, and chondrogenic lineages. The effectiveness of these methods hinges on the precise combination and concentration of inducing agents, which activate specific and often competitive signaling cascades within the cell. A deep understanding of these underlying mechanisms—such as the pivotal role of ERK activation in promoting osteogenesis over adipogenesis—is not merely academic but fundamentally enhances our ability to design robust experiments and develop novel therapeutic strategies for a range of degenerative diseases and injuries. As the field progresses, these established protocols will continue to serve as a critical benchmark against which newer technologies, such as the manipulation of epigenetic regulators or RNA storage systems like P-bodies, can be evaluated and integrated [46]. Adherence to these detailed protocols, coupled with rigorous validation, will ensure the generation of high-quality, reproducible data that advances both basic stem cell biology and clinical translation in regenerative medicine.
The directed differentiation of stem cells into specific lineages represents a cornerstone of regenerative medicine. Small molecules, with their cell-permeable nature, cost-effectiveness, and reversible activity, have emerged as powerful tools to precisely manipulate stem cell fate [47] [48]. Among these, statins, metformin, and adenosine have demonstrated significant potential as inducers of osteogenic, chondrogenic, and adipogenic differentiation. Their ability to activate or inhibit key intracellular signaling pathways allows for the controlled specification of mesenchymal stem cells (MSCs) into desired lineages, offering substantial promise for tissue engineering and therapeutic applications [47] [49]. This Application Note provides a detailed overview of the mechanisms, optimal concentrations, and experimental protocols for using these small molecules to direct stem cell differentiation, serving as a practical guide for researchers and drug development professionals.
Small Molecule Mechanisms and Signaling Pathways
Key Signaling Pathways in Stem Cell Differentiation
Small molecules guide stem cell fate by modulating specific signaling pathways. The table below summarizes the primary pathways involved in osteogenic, chondrogenic, and adipogenic differentiation.
Table 1: Key Signaling Pathways in Stem Cell Differentiation
| Pathway | Pro-Differentiation Role | Key Effectors | Small Molecule Modulators |
|---|---|---|---|
| BMP/Smad | Pro-osteogenic, Pro-adipogenic [48] | Smad1/5/8, Smad4, BMP-2 [48] [50] | Statins, Metformin [47] [49] |
| Wnt/β-catenin | Pro-osteogenic [48] | β-catenin, GSK-3, LRP5/6 [48] | Metformin, Icarin [47] |
| Adenosine Signaling | Pro-osteogenic, Anti-adipogenic [48] | A2b receptor [51] [48] | Adenosine |
| Hedgehog | Pro-osteogenic, Anti-adipogenic [48] | Smoothened, Gli transcription factors [48] | Atractylenolides [47] |
| AMPK | Pro-osteogenic (context-dependent) [52] | AMPK | Metformin, Arctigenin [47] [52] |
| RhoA-ROCK | Anti-chondrogenic (inhibition promotes) [50] | RhoA, ROCK | Fluvastatin [50] |
Visualizing Key Signaling Pathways
The following diagram illustrates the core signaling pathways through which statins, metformin, and adenosine exert their effects on stem cell differentiation.
Statins as Differentiation Inducers
Mechanism of Action
Statins, primarily known as cholesterol-lowering drugs, inhibit 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA) reductase, the rate-limiting enzyme in the mevalonate pathway [53] [49]. This inhibition depletes downstream metabolites like geranylgeranyl pyrophosphate (GGPP), essential for prenylation of small GTP-binding proteins such as RhoA [50]. The subsequent inhibition of RhoA-ROCK signaling upregulates bone morphogenetic protein 2 (BMP-2) expression, a critical driver of osteogenic and chondrogenic differentiation [50] [49]. Lipophilic statins (e.g., simvastatin, fluvastatin) are particularly effective due to superior cell membrane permeability [53] [49].
Application Notes and Experimental Protocols
Osteogenic Differentiation of MSCs
- Optimal Concentrations:
- Fluvastatin: 0.1 - 1 μM significantly increased bone volume fraction, trabecular organization, and expression of osteogenic markers (RUNX2, SPP1, COL1A2) in an embryonic chick femur model [53].
- Simvastatin: 0.1 - 1 μM demonstrated similar osteoinductive effects. Doses of 10 μM attenuated these beneficial outcomes, indicating a narrow therapeutic window [53].
- Cell Sources: Bone marrow-derived MSCs (BM-MSCs), adipose-derived MSCs (AD-MSCs), and organotypic bone models [53] [49].
- Key Readouts:
Chondrogenic Differentiation of AD-MSCs
- Optimal Concentration: 0.1 μM fluvastatin in pellet culture without exogenous growth factors [50].
- Protocol:
- Cell Culture: Culture human AD-MSCs (hADMSCs) in high-glucose DMEM supplemented with 1% FBS [50].
- Pellet Formation: Seed 2 × 10^5 cells/well in 96-well U-bottom plates. Centrifuge at 300-500 × g for 5 minutes and incubate for 48 hours to form 3D pellets [50].
- Chondrogenic Induction: Culture pellets for 2-3 weeks in chondrogenic medium (high-glucose DMEM, 1% ITS, 40 μg/mL L-proline, 0.1 μM dexamethasone, 50 μg/mL L-ascorbic acid, 110 μg/mL sodium pyruvate) supplemented with 0.1 μM fluvastatin. Replace media every 3-4 days [50].
- Inhibition Studies: To confirm mechanism, add 100 μM mevalonic acid (MVA) or 20 μM GGPP to the chondrogenic medium with fluvastatin. Alternatively, use 20 μM Y27632 (ROCK inhibitor) to mimic fluvastatin's effect [50].
- Key Readouts:
Table 2: Summary of Statin Effects on Stem Cell Differentiation
| Statin | Target Lineage | Optimal Concentration | Key Upregulated Markers | Experimental Model |
|---|---|---|---|---|
| Fluvastatin | Osteogenic [53] | 0.1 - 1 μM | RUNX2, SPP1, COL1A2 [53] | Embryonic chick femur ex vivo [53] |
| Simvastatin | Osteogenic [53] | 0.1 - 1 μM | RUNX2, SPP1, COL1A2 [53] | Embryonic chick femur ex vivo [53] |
| Fluvastatin | Chondrogenic [50] | 0.1 μM | BMP2, SOX9, ACAN, COL2A1 [50] | Human AD-MSC pellet culture [50] |
Metformin as a Bifunctional Differentiation Inducer
Mechanism of Action
Metformin, a first-line antidiabetic drug, exerts its effects primarily through activation of AMP-activated protein kinase (AMPK) [52]. This activation governs cross-talk with other pathways, including Akt and BMP, leading to context-dependent outcomes on stem cell fate. Notably, metformin can induce both osteogenic and adipogenic differentiation, with the resulting lineage commitment highly dependent on cell source, microenvironment, and concentration [52] [54].
Application Notes and Experimental Protocols
Osteogenic Differentiation of ASCs
- Optimal Concentration: 500 μM in culture medium [52].
- Cell Source: Rat adipose-derived stem cells (rASCs) [52].
- Protocol:
- Cell Seeding: Plate rASCs at a standard density (e.g., 5,000-10,000 cells/cm²) in complete growth medium.
- Osteogenic Induction: Upon reaching 70-80% confluence, switch to osteogenic induction medium (OM). A standard OM consists of:
- High-glucose DMEM
- 10% Fetal Bovine Serum (FBS)
- 10 mM β-glycerophosphate
- 50 μM L-ascorbic acid
- 0.1 μM dexamethasone
- 500 μM metformin [52].
- Culture Duration: Maintain cultures for 14-21 days, changing the medium every 2-3 days.
- Key Readouts:
- Early Markers: Alkaline phosphatase (ALP) activity after 7-10 days [52].
- Late Markers: Extracellular matrix mineralization (von Kossa or Alizarin Red S staining), osteocalcin secretion (ELISA or immunostaining) after 21-28 days [52].
- Gene Expression: qRT-PCR for Runx2, Alp, Osteocalcin, and Collagen I [52].
Adipogenic Differentiation of UC-MSCs
- Optimal Concentration: 3 mM in culture medium [54].
- Cell Source: Human umbilical cord MSCs (UC-MSCs) [54].
- Protocol:
- Cell Seeding: Plate UC-MSCs in standard growth medium.
- Adipogenic Induction: At confluence, induce adipogenesis using a commercial kit or standard adipogenic medium supplemented with 3 mM metformin. The medium is typically replaced every 3-4 days.
- Culture Duration: 14-21 days.
- Key Readouts:
Table 3: Summary of Metformin's Bifunctional Effects on Stem Cells
| Cell Source | Induced Lineage | Metformin Concentration | Key Molecular Effects | Primary Readouts |
|---|---|---|---|---|
| Rat ASCs [52] | Osteogenic | 500 μM | Activates AMPK and BMP-2 [52] | ↑ ALP activity, ↑ Mineralization (Von Kossa), ↑ Osteocalcin [52] |
| Human UC-MSCs [54] | Adipogenic | 3 mM | ↑ PPARγ, ↓ FABP4, Anti-inflammatory (↓ IL-6, MCP-1) [54] | ↑ Lipid droplets (Oil Red O), Immunomodulation [54] |
Adenosine as an Osteogenic Inducer
Mechanism of Action
Adenosine, a purine nucleoside, promotes osteogenic differentiation primarily through the A2b adenosine receptor (A2bR), a Gs/q-protein-coupled receptor [51] [48]. Activation of A2bR signaling upregulates genes associated with osteogenesis and increases osteocalcin protein expression [51]. Studies on A2bR knockout mice confirm its critical role, showing decreased osteogenic potential of MSCs, lower bone density, and delayed fracture repair [51].
Application Notes and Experimental Protocols
Osteogenic Differentiation of Pluripotent Stem Cells
- Induction Method: Culture of hESCs on biomineralized calcium phosphate (CaP) matrices, which naturally release phosphate ions, triggering endogenous adenosine signaling and A2bR activation [51]. Exogenous adenosine supplementation is an alternative approach.
- Cell Source: Human embryonic stem cells (hESCs), human-induced pluripotent stem cells (hiPSCs), human MSCs (hMSCs) [51].
- Protocol for Biomaterial-Mediated Induction:
- Matrix Preparation: Fabricate and mineralize synthetic matrices, such as PEGDA-co-A6ACA hydrogels, in modified simulated body fluid (m-SBF) to form osteoinductive CaP coatings [51].
- Cell Seeding: Seed hESCs or hiPSCs onto the mineralized matrices in standard maintenance medium without osteogenic supplements.
- Culture Duration: Maintain cultures for up to 28 days to observe spontaneous osteogenic differentiation.
- Inhibition Control: To confirm the role of A2bR, include a group with an A2bR-specific antagonist (e.g., PSB 603) [51].
- Key Readouts:
- Gene Expression: qRT-PCR for early and late osteogenic markers (e.g., Runx2, Osterix, Osteocalcin).
- Protein Expression: Immunostaining for Osteocalcin (OCN) [51].
- Functional Assay: Alkaline phosphatase activity.
The Scientist's Toolkit: Essential Research Reagents
The following table lists key reagents used in the cited studies for inducing differentiation with statins, metformin, and adenosine.
Table 4: Essential Research Reagents for Differentiation Studies
| Reagent / Tool | Function / Application | Example Usage in Protocols |
|---|---|---|
| Fluvastatin [53] [50] | HMG-CoA reductase inhibitor; induces osteo/chondrogenesis via BMP-2. | 0.1 μM in chondrogenic pellet culture of hADMSCs [50]. |
| Simvastatin [53] [49] | Lipophilic HMG-CoA reductase inhibitor; promotes osteogenesis. | 0.1 - 1 μM in ex vivo bone models [53]. |
| Metformin [52] [54] | AMPK activator; induces osteogenic or adipogenic differentiation. | 500 μM for rASC osteogenesis; 3 mM for UC-MSC adipogenesis [52] [54]. |
| Mevalonic Acid (MVA) [50] | Metabolite downstream of HMG-CoA; rescues statin effects. | 100 μM to confirm statin mechanism via mevalonate pathway [50]. |
| Geranylgeranyl Pyrophosphate (GGPP) [50] | Isoprenoid for protein prenylation; rescues statin effects. | 20 μM to confirm role of small GTPase inhibition in statin action [50]. |
| Y27632 (ROCK Inhibitor) [50] | Inhibits ROCK kinase; mimics statin effect on chondrogenesis. | 20 μM in control chondrogenic medium to promote differentiation [50]. |
| Noggin [50] | BMP antagonist; inhibits BMP-mediated differentiation. | 500 ng/mL to block statin-induced chondrogenesis [50]. |
| A2bR Antagonist (e.g., PSB 603) [51] | Inhibits A2b adenosine receptor; confirms adenosine signaling role. | Used in culture to attenuate CaP matrix-driven osteogenesis [51]. |
| Biomineralized CaP Matrices [51] | Osteoinductive biomaterial; activates endogenous adenosine signaling. | Scaffold for hESC culture to drive osteogenesis without soluble inducers [51]. |
Statins, metformin, and adenosine are potent inducers of stem cell differentiation, each acting through distinct and well-characterized signaling pathways. The efficacy of these small molecules is profoundly influenced by cell type, concentration, and culture environment. Fluvastatin and simvastatin at low concentrations (0.1-1 μM) are robust inducers of osteogenic and chondrogenic commitment, primarily via BMP-2 upregulation. Metformin exhibits a unique bifunctionality, promoting osteogenesis in ASCs at 500 μM and adipogenesis in UC-MSCs at 3 mM. Adenosine signaling, particularly through the A2b receptor, serves as a critical mediator of osteogenesis, especially in the context of biomaterial-based strategies. The protocols and data summarized herein provide a foundational framework for researchers aiming to harness these small molecules for advanced tissue engineering and regenerative medicine applications.
The field of regenerative medicine is increasingly focused on developing advanced biomaterial scaffolds that not only provide structural support but also actively direct stem cell fate. Within the context of a broader thesis on stem cell differentiation into osteogenic, chondrogenic, and adipogenic lineages, this document details the significant role of synthetic acrylate-based polymers functionalized with natural extracellular matrix (ECM) components, specifically gelatin and heparin. These functionalized scaffolds demonstrate a profound ability to influence mesenchymal stem cell (MSC) commitment and differentiation, offering promising pathways for bone and cartilage tissue engineering [55].
Acrylate-based polymers provide a highly customizable and biocompatible foundation, allowing for precise manipulation of mechanical properties and chemical functionality. The grafting of gelatin, a denatured collagen that retains Arg-Gly-Asp (RGD) sequences, enhances cell adhesion and survival [55] [56]. Conversely, heparin, a sulfated glycosaminoglycan, possesses a strong affinity for a wide range of growth factors and ECM proteins, stabilizing them and presenting them to cells to direct differentiation processes [55] [57]. The combination of these materials creates a powerful platform for controlling the stem cell microenvironment, thereby influencing lineage specification and loss of multipotency.
Impact on Stem Cell Differentiation
Osteogenic Differentiation
Research indicates that acrylate-based scaffolds functionalized with specific chemical groups and biomolecules can significantly promote osteogenic differentiation. A key study on human bone marrow MSCs (hBMMSCs) cultured on acrylate substrates with grafted heparin showed spontaneous osteogenic commitment even in basal medium conditions, without the need for exogenous osteoinductive factors [55]. This was further enhanced when cultures were maintained in osteogenic medium for 21 days. The heparin functionalization is believed to promote osteogenesis by inducing an extended conformation of adsorbed fibronectin, which in turn mediates favorable cell-biomaterial interactions [55].
Independent research using plasma polymerisation to deposit specific chemical groups on polymer scaffolds found that amine (NH₂) group functionalization significantly enhanced osteogenic differentiation of human adipose-derived stem cells (ADSCs). This was evidenced by the upregulated gene and protein expression of classic osteogenic markers, including collagen I, alkaline phosphatase, and osteocalcin [58].
Chondrogenic Differentiation
The same study on chemical group functionalization revealed that carboxyl (COOH) groups preferentially direct ADSC differentiation toward the chondrogenic lineage, marked by increased expression of aggrecan and collagen II [58]. Furthermore, three-dimensional environments created from gelatin and heparin have proven highly effective for cartilage tissue engineering. Biofabricated hydrogels from allylated gelatin (GelAGE) and thiolated heparin (HepSH) support robust chondrogenesis [57]. The incorporation of HepSH within the gelatin matrix acts as a biological amplifier, enhancing the cellular response to other cues such as matrix stiffness and oxygen availability, thereby facilitating the development of more physiologically relevant, zonal cartilage models [57].
Maintenance and Loss of Multipotency
A critical finding is that the interaction of MSCs with grafted biomolecules can itself induce lineage commitment and a consequent loss of multipotency, even in the absence of specific differentiation media [55]. This underscores the powerful influence of the substrate's biochemical composition. Factors such as substrate topography, surface charge, and microstructure have also been demonstrated to significantly impact the expression of multipotency markers in hBMMSCs [55]. Therefore, careful design of the scaffold is essential, whether the goal is to maintain multipotency during cell expansion or to direct differentiation toward a specific lineage.
The following tables summarize key quantitative findings from recent studies on the effects of biomaterial scaffolds on stem cell behavior.
Table 1: Impact of Scaffold Composition on Stem Cell Differentiation Markers
| Scaffold Type | Biomolecule/Chemical Group | Cell Type | Key Findings (Gene/Protein Expression) | Reference |
|---|---|---|---|---|
| Poly(EA-co-EMA-co-AAc) Film | Heparin (Grafted) | hBMMSCs | Spontaneous osteogenic commitment in basal medium; enhanced differentiation in osteogenic medium. | [55] |
| Plasma Polymerised Scaffold | Amine (NH₂) | ADSCs | ↑ Osteocalcin, ↑ Alkaline Phosphatase, ↑ Collagen I (Gene & Protein Level) | [58] |
| Plasma Polymerised Scaffold | Carboxyl (COOH) | ADSCs | ↑ Aggrecan, ↑ Collagen II (Gene Level) | [58] |
| GelAGE-HepSH Hydrogel | Heparin (Covalent) | Chondrocytes | Enhanced & more uniform ECM secretion; amplified response to stiffness/oxygen cues. | [57] |
Table 2: Effect of GelAGE-HepSH Hydrogel Stiffness on Chondrogenesis
| Hydrogel Stiffness | Impact on Cell Localization & Tissue Formation | Impact on ECM Secretion | Suitability for Cartilage Models |
|---|---|---|---|
| Soft (12 kPa) | Irregular cell localization | Not specified in results | Low |
| Medium (55 kPa) | Uniform tissue formation; maintained shape fidelity | Uniform | High |
| Stiff (121 kPa) | Restricted overall tissue formation | Restricted | Low |
Experimental Protocols
Protocol 1: Fabrication of Acrylate-Based Films with Grafted Gelatin or Heparin
This protocol describes the synthesis of flat acrylate copolymer films and the subsequent covalent grafting of biomolecules, adapted from a study investigating hBMMSC behavior [55].
1. Synthesis of Poly(EA-co-EMA-co-AAc) Copolymers: - Reagents: Ethyl Acrylate (EA), Ethyl Methacrylate (EMA), Acrylic Acid (AAc), acetone (solvent), benzoin (photoinitiator). - Procedure: a. Mix monomers EA, EMA, and AAc in desired ratios (e.g., 45:45:10 for 10 wt% AAc) with 30 wt% acetone and 0.5 wt% benzoin. b. Transfer the mixture to a transparent mould and expose to UV light (e.g., TL 05-8W Philips lamp) for 24 hours. c. Perform a final thermal treatment at 90°C overnight to ensure full monomer conversion and evaporate acetone. d. Purify the resulting block copolymer by successive dissolution in acetone (10% w/v) and precipitation in ultrapure water. e. Dry the purified polymer in a vacuum chamber at 25°C for 48 hours and store in a dry atmosphere.
2. Preparation of Flat Films via Solvent Casting: - Prepare a 6% (wt/wt) solution of the purified copolymer in acetone under continuous stirring for 24 hours. - Pour the solution onto a Teflon plate and allow the solvent to evaporate under a fume hood at 25°C for 24 hours. - Dry the films further in a vacuum oven at 50°C for 3 days to remove any residual solvent.
3. Surface Functionalization with Gelatin or Heparin: - Grafting: Covalently graft gelatin or heparin onto the surface of the acrylate films (AAc5 and AAc10, containing carboxylic acid groups) using standard carbodiimide chemistry (e.g., using EDC/NHS). The specific detailed reaction steps for this particular system are available in the supplementary information of the source material [55].
Protocol 2: Biofabrication of Gelatin-Heparin (GelAGE-HepSH) Hydrogels for 3D Chondrogenesis
This protocol outlines the creation of a photopolymerizable hydrogel system for engineering zonal cartilage models [57].
1. Material Synthesis: - Allylated Gelatin (GelAGE): Synthesize by reacting gelatin from porcine skin with allyl glycidyl ether (AGE) under basic conditions. Purify via dialysis and lyophilize. - Thiolated Heparin (HepSH): Synthesize by reacting heparin sodium salt with cysteamine using carbodiimide chemistry (EDC/HOBt). Purify via dialysis and lyophilize.
2. Hydrogel Precursor Preparation: - Prepare a sterile precursor solution by dissolving GelAGE and HepSH in a suitable cell-compatible buffer (e.g., PBS). A typical concentration is 6% (w/v) GelAGE and 1% (w/v) HepSH. - Add a photoinitiator (e.g., Irgacure 2959) to a final concentration of 0.1% (w/v).
3. 3D Cell Encapsulation and Crosslinking: - Mix the hydrogel precursor solution with a chondrocyte or MSC suspension to achieve the desired final cell density (e.g., 5-10 million cells/mL). - Pipette the cell-laden solution into a mould of the desired geometry (e.g., sphere, disc) or load into a bioprinter for more complex architectures. - Expose the construct to UV light (e.g., 365 nm, 5-10 mW/cm²) for 60-120 seconds to initiate crosslinking via a thiol-ene reaction between the allyl and thiol groups.
4. Culture and Chondrogenic Induction: - Culture the fabricated hydrogels in chondrogenic medium (e.g., DMEM high glucose, supplemented with ITS+1, L-ascorbic acid-2-phosphate, dexamethasone, proline, and TGF-β3). - Maintain cultures for up to 4-6 weeks, changing the medium 2-3 times per week, to allow for cartilage-like tissue development.
Signaling Pathways and Experimental Workflow
The following diagrams illustrate the key signaling mechanisms influenced by these biomaterials and a generalized workflow for creating and testing the scaffolds.
Biomaterial-Mediated Stem Cell Fate
Biomaterial Scaffold Workflow
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Materials for Acrylate-Gelatin-Heparin Scaffold Research
| Reagent/Material | Function/Description | Key Role in Research |
|---|---|---|
| Ethyl Acrylate (EA) / Ethyl Methacrylate (EMA) | Methacrylate monomers forming the polymer backbone. | Provides the primary, synthetic structural component of the scaffold with tunable mechanical properties. |
| Acrylic Acid (AAc) | A functional monomer containing a carboxylic acid group. | Introduces reactive carboxyl groups onto the polymer backbone for subsequent covalent grafting of biomolecules. |
| Gelatin (Type A) | Denatured collagen derived from porcine skin. | Provides bioadhesive RGD sequences to enhance cell adhesion, migration, and survival. |
| Heparin Sodium Salt | Sulfated glycosaminoglycan from porcine intestinal mucosa. | Binds and stabilizes a wide array of growth factors (e.g., BMP, TGF-β), modulating their presentation to cells to direct differentiation. |
| 1-Ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC) | Carbodiimide crosslinker. | Activates carboxylic acid groups for covalent amide bond formation with primary amines on gelatin or during heparin thiolation. |
| Photoinitiator (e.g., Benzoin, Irgacure 2959) | Compound that generates reactive species upon UV light exposure. | Initiates the free-radical polymerization of acrylate monomers or the crosslinking of modified polymers like GelAGE. |
| Allyl Glycidyl Ether (AGE) | Ether compound used for chemical modification. | Used to synthesize allylated gelatin (GelAGE) by introducing photopolymerizable allyl groups. |
| Cysteamine | Aminothiol compound. | Used to synthesize thiolated heparin (HepSH) by introducing free thiol groups for crosslinking via thiol-ene reaction. |
The pursuit of directing stem cell differentiation into osteogenic, chondrogenic, and adipogenic lineages has moved beyond two-dimensional culture into the third dimension. Advanced 3D environments—utilizing bioprinting, microspheres, and decellularized extracellular matrix (dECM)—are revolutionizing this field by providing biomimetic niches that closely replicate the structural, biochemical, and mechanical cues of native tissue. These platforms offer unprecedented control over the stem cell microenvironment, enabling more precise investigation of differentiation mechanisms and the development of robust, clinically applicable tissue-engineered constructs. This document provides detailed application notes and standardized protocols for implementing these technologies within a stem cell differentiation research framework.
Research Reagent Solutions
The following table catalogues essential materials and reagents critical for fabricating and utilizing advanced 3D environments for stem cell research.
Table 1: Key Research Reagents and Materials for 3D Stem Cell Environments
| Reagent/Material | Function/Application | Specific Examples & Notes |
|---|---|---|
| Decellularized ECM (dECM) | Provides tissue-specific biochemical cues; used as a scaffold or bioink component. | Porcine Small Intestine Submucosa (SIS) [59]; Bone-derived dECM [60]; Retinal-derived dECM (RdECM) [61]. |
| Structural Proteins | Core scaffold components providing mechanical integrity and cell adhesion sites. | Collagen I (C), Collagen IV (CIV), Laminin 411 (LN411), Fibronectin (FN) [62]. |
| Synthetic Polymers | Enhances mechanical properties and printability of bioinks. | Polycaprolactone (PCL) [63]; Alginate (often ionically crosslinked with CaCl₂) [60]. |
| Photo-initiators | Enables light-based crosslinking (photopolymerization) of hydrogels during bioprinting. | Lithium phenyl-2,4,6-trimethylbenzoylphosphinate (LAP), Irgacure 2959 [60]. |
| Growth Factors | Soluble signaling molecules that direct stem cell fate. | Bone Morphogenic Protein-2 (BMP-2) for osteogenesis [60]; Vascular Endothelial Growth Factor (VEGF) for endothelial differentiation [62] [64]. |
| Detergents & Enzymes | Agents for tissue decellularization. | Ionic: Sodium Dodecyl Sulfate (SDS); Non-ionic: Triton X-100; Enzymatic: Trypsin, DNase [64] [63] [61]. |
Application Notes
Optimizing the Extracellular Matrix for Enhanced Differentiation
The specific composition of the ECM is a powerful determinant of stem cell fate. A Design of Experiments (DoE) approach has been successfully used to move beyond single-protein substrates and optimize complex ECM formulations. For instance, a combination of Collagen I, Collagen IV, and Laminin 411 was identified as a potent inducer of endothelial differentiation, outperforming the commonly used Matrigel [62]. The DoE methodology systematically evaluated protein interactions and identified key signaling axes regulated by ECM stimulation, providing a blueprint for optimizing microenvironments for other lineages like osteogenesis and chondrogenesis [62].
A critical finding is that the role of ECM components can be context-dependent. While Fibronectin (FN) was essential for initial cell attachment, its subsequent removal from the optimized "TheO" formulation resulted in a significantly more potent "Endothelial Optimized" (EO) matrix (TheO-FN). This highlights that attachment-supporting factors are not always synonymous with differentiation-promoting factors, a crucial consideration when designing differentiation protocols [62]. Furthermore, the ECM acts as a reservoir for growth factors; pre-incubating the ECM with VEGF before cell seeding enhanced differentiation outcomes by capitalizing on this sequestration capacity [62].
3D Bioprinting for Spatially Defined Differentiation
3D bioprinting enables the precise spatial organization of cellular niches, biochemical signals, and mechanical properties within a single construct. This is particularly valuable for engineering complex tissue interfaces, such as the osteochondral junction, which requires a seamless gradient from bone to cartilage [65].
- Bioprinting Modalities: The primary bioprinting approaches include inkjet, extrusion, laser-assisted, and stereolithography-based methods. Extrusion bioprinting is widely used for its ability to handle high-cell-density bioinks and a wide range of materials [65].
- Bioink Innovations: A major trend is the use of dECM-based bioinks, which preserve tissue-specific biochemical signals. For example, a retina-on-a-chip was fabricated using a hybrid bioink of retinal-derived dECM (RdECM) and vascular-derived dECM (VdECM) to create a biomimetic microenvironment [61]. For bone tissue engineering, natural polymer hydrogels like gelatin-methacryloyl (GelMA) are favored for their osteoid-mimicry and tunable mechanical properties [60].
- Crosslinking Chemistry: The choice between chain-growth (e.g., GelMA) and step-growth crosslinking is critical. It dictates the network structure, swelling, degradation rate, and ultimately, how cells experience their microenvironment, influencing processes like migration and differentiation [60].
dECM Microspheres and Spheroids for High-Throughput Screening
MatriSpheres represent a hydrogel-free, self-assembly platform for establishing ECM-rich 3D models. In this approach, solubilized dECM (e.g., from small intestine submucosa) is organized by cells into intercellular, stroma-like regions within spheroids over just five days [59]. This method is distinct from passive hydrogel embedding and promotes cell-driven tissue morphogenesis.
The key advantage for drug development is that these MatriSpheres exhibit ECM-dependent transcriptional and cytokine profiles associated with disease states and metabolism. Model benchmarking has shown that MatriSpheres enhance correlation with in vivo tumor cells compared to traditional ECM-poor spheroids, making them a high-fidelity tool for disease modeling and high-throughput drug screening [59]. This platform can be adapted to model stem cell niches in health and disease for more predictive therapeutic evaluation.
Experimental Protocols
Protocol: DoE-Optimized ECM Coating for Enhanced Differentiation
This protocol outlines the procedure for creating and testing an optimized ECM coating to drive stem cell differentiation, based on a validated DoE approach [62].
Table 2: Optimized ECM Formulation for Enhanced Differentiation
| ECM Component | Optimized Concentration | Significance/Effect |
|---|---|---|
| Collagen I (C) | 35.6 µg/mL | Positive but small effect size on differentiation. |
| Collagen IV (CIV) | 67.2 µg/mL | Large positive effect size; critical for differentiation. |
| Laminin 411 (LN411) | 0.9 µg/mL | Large positive effect size; key signaling component. |
| Fibronectin (FN) | 0 µg/mL (removed) | Required for cell attachment, but inhibited maximal differentiation. |
Materials:
- Purified proteins: Collagen I, Collagen IV, Laminin 411, Fibronectin.
- Sterile phosphate-buffered saline (PBS).
- Cell culture plates.
- Human induced pluripotent stem cells (hiPSCs).
Procedure:
- Solution Preparation: Prepare a sterile stock solution of the optimized ECM (EO) formulation in PBS, containing 35.6 µg/mL Collagen I, 67.2 µg/mL Collagen IV, and 0.9 µg/mL Laminin 411. Note: Fibronectin is omitted from the final differentiation coating.
- Surface Coating: Add enough ECM solution to cover the surface of the culture vessel. Incubate for at least 1 hour at room temperature or overnight at 4°C.
- Pre-incubation with Growth Factors (Optional): To enhance differentiation, add a solution of VEGF (e.g., 10 ng/mL in PBS) to the coated wells. Incubate for 30 minutes at room temperature, then aspirate. [62]
- Cell Seeding and Differentiation: Aspirate any remaining coating solution. Seed hiPSCs at the desired density and initiate the specific differentiation protocol (e.g., by adding CHIR99021 for endothelial differentiation [62]).
- Validation: Assess differentiation efficiency via immunostaining for lineage-specific markers (e.g., CD31 for endothelial cells) and quantitative PCR.
Protocol: Fabrication of ECM-Rich MatriSpheres
This protocol describes the generation of 3D MatriSpheres using decellularized ECM to create a high-fidelity microenvironment for stem cell culture and drug testing [59].
Materials:
- Solubilized dECM (e.g., from porcine small intestine submucosa, pepsin-digested).
- Target cells (e.g., stem cells, primary tissue cells).
- Standard cell culture medium.
- Low-attachment 96-well or 384-well spherical plate.
- Centrifuge.
Procedure:
- dECM Preparation: Thaw solubilized dECM on ice. Keep it on ice at all times to prevent premature gelation.
- Cell-ECM Mixture: Create a single-cell suspension and mix it with the solubilized dECM at a sub-gelation concentration on ice. The final ECM concentration will depend on the tissue source but is typically between 1-4 mg/mL. The cell density should be optimized for the specific application.
- Sphere Formation: Pipette the cell-ECM mixture into the wells of a low-attachment round-bottom plate.
- Centrifugation: Centrifuge the plate at a low speed (e.g., 300-500 x g for 3-5 minutes) to pellet the cells and ECM mixture at the bottom of the well, initiating aggregation.
- Culture and Maturation: Incubate the plate at 37°C, 5% CO₂ for up to 5 days. During this time, the cells will actively organize the dECM into intercellular stroma-like regions.
- Application: Use the matured MatriSpheres for downstream assays, such as transcriptional analysis, cytokine profiling, or high-throughput drug screening [59].
Protocol: Decellularization of Tissues for dECM Bioink
This is a generalized protocol for tissue decellularization, adaptable to various source tissues (e.g., retina, tendon, bone) for the production of dECM bioinks [64] [63] [61].
Materials:
- Source tissue (e.g., porcine retina, small intestine).
- Deionized Water (DW).
- Phosphate-Buffered Saline (PBS).
- Chemical agents: SDS, Triton X-100, Ethylenediaminetetraacetic acid (EDTA).
- Enzymatic agents: DNase.
- Physical equipment: Shaker, freezer (-80°C).
Procedure:
- Initial Wash and Preparation: Rinse the isolated tissue thoroughly with DW and then with PBS for 24 hours to remove residual blood and impurities [61].
- Physical Disruption (Optional): Perform freeze-thaw cycles (e.g., between -80°C and 37°C) to lyse cell membranes via ice crystal formation. This is often used as a first step [63].
- Chemical Treatment:
- Treat tissue with an ionic detergent (e.g., 0.1% SDS) for several hours to solubilize cell membranes and nuclear components [61].
- Wash extensively with PBS to remove the detergent.
- Treat with a non-ionic detergent (e.g., 2% Triton X-100) often combined with a chelating agent (e.g., 25 mM EDTA) for a longer duration (e.g., 72 hours) to remove remaining cellular material [61].
- Wash again with PBS.
- Enzymatic Treatment: Incubate the tissue with DNase (e.g., 100 U/mL) to digest residual DNA [61].
- Final Washes and Storage: Perform a final series of PBS washes. The resulting acellular dECM can be lyophilized and milled into a powder for long-term storage or digested (e.g., with pepsin) to create a solubilized dECM bioink [59] [61].
Signaling and Workflow Diagrams
Workflow for Creating 3D Stem Cell Microenvironments
Signaling Pathways in 3D Microenvironment-Driven Differentiation
The therapeutic potential of stem cells in regenerative medicine is vast, hinging on their ability to differentiate into specific cell lineages such as osteogenic (bone), chondrogenic (cartilage), and adipogenic (fat) cells. A significant challenge in clinical applications is the unpredictability of differentiation outcomes, which are influenced by donor variability, culture conditions, and cellular heterogeneity. Traditional methods for assessing differentiation potential are often destructive, time-consuming, and provide only endpoint measurements. The integration of quantitative morphological analysis with machine learning (ML) is revolutionizing this field by enabling non-destructive, early, and accurate prediction of stem cell fate. This Application Note details the protocols and data analysis frameworks that leverage cellular morphology, captured via simple imaging techniques, to forecast differentiation outcomes long before conventional markers are detectable.
Key Morphological Features Predictive of Differentiation
Cellular morphology is a direct reflection of a cell's functional state and differentiation commitment. The following table summarizes key morphological features that have been correlated with specific differentiation lineages.
Table 1: Morphological Features Predictive of Differentiation Lineage
| Differentiation Lineage | Key Morphological Features | Relationship to Differentiation Potential |
|---|---|---|
| Osteogenic | Increased Cell Area | Higher cell spreading area correlates with osteogenic commitment [66] [67]. |
| High Aspect Ratio | Elongated, spindle-like shapes are associated with osteogenic potential [67]. | |
| Increased Edge Roughness | Complex cell boundaries are predictive of successful bone cell differentiation [67]. | |
| Adipogenic | Low Cell Area | Rounded cells with limited spreading are indicative of adipogenic fate [66]. |
| Low Aspect Ratio | A more rounded, bulky cell morphology is a hallmark of adipogenic differentiation [66]. | |
| Chondrogenic | (Information not explicitly covered in search results) | |
| Pluripotent State | Large Nucleus, Scant Cytoplasm | Features of undifferentiated hESCs include a large nuclear-to-cytoplasmic ratio [68]. |
| Short Intercellular Distance | Large, dense colonies with minimal gaps between cells are characteristic of pluripotency [68]. |
Machine Learning Models for Morphological Prediction
Various machine learning models, particularly deep learning-based convolutional neural networks (CNNs), have been successfully applied to predict differentiation from cell images. The performance of different models varies based on their architecture and the dataset.
Table 2: Comparison of Machine Learning Models for Differentiation Prediction
| Model Name | Best For | Key Advantages | Reported Performance | Limitations & Considerations |
|---|---|---|---|---|
| ResNet-50 | Osteogenic & Adipogenic prediction from high-res images [67] | Residual blocks prevent vanishing gradient problem in deep networks [67]. | AUC > 0.96, Accuracy up to 96.3% for early (24h) osteogenic prediction [67]. | Requires large datasets (>10,000 images); sensitive to imaging parameters and label noise [67]. |
| Vision Transformers (ViTs) | General cell image analysis | Self-attention mechanism captures global cellular features effectively [67]. | Promising for medical image recognition; performance data in this specific context is emerging [67]. | Computational complexity and data requirements can be high [67]. |
| Random Forest | Early prediction of muscle stem cell (MuSC) efficiency [69] | Effective with hand-crafted features (e.g., FFT-based vectors); less computationally intensive [69]. | Enabled prediction of MuSC efficiency ~50 days before endpoint using phase-contrast images [69]. | Performance dependent on quality of feature extraction method. |
| VGG19 | Benchmarking | Established architecture | Lower performance (AUC ~0.89) and prone to overfitting on small datasets compared to ResNet-50 [67]. | Architecture is computationally expensive relative to performance [67]. |
| InceptionV3 | Benchmarking | Multi-scale feature extraction | Lower performance (AUC = 0.89) in classifying cell morphology [67]. | May not be optimal for cell shape classification tasks [67]. |
Detailed Experimental Protocols
Protocol: Early Prediction of Osteogenic Differentiation using ResNet-50
This protocol outlines the steps for predicting the osteogenic differentiation potential of human Mesenchymal Stem Cells (hMSCs) from bright-field images using a ResNet-50 model [67].
Key Reagents:
- Cells: Human Mesenchymal Stem Cells (hMSCs).
- Culture Medium: Standard growth medium for hMSCs.
- Osteogenic Induction Medium: Typically contains dexamethasone, β-glycerolphosphate, and ascorbate-2-phosphate [70] [71].
- Imaging Equipment: Phase-contrast or bright-field microscope.
Procedure:
- Cell Seeding and Culture: Seed hMSCs at an appropriate density in multi-well plates. Maintain cells in growth medium until they reach the desired confluence.
- Image Acquisition: At defined time points (e.g., days 0, 1, 3, 5, and 7 post-induction), acquire bright-field images of the cells using a microscope. Ensure consistent imaging parameters (e.g., resolution, lighting) across all samples.
- Data Labeling (for training): Upon completion of the culture, assess the osteogenic differentiation efficiency of each sample using a standard endpoint assay, such as Alkaline Phosphatase (ALP) activity or Alizarin Red S staining for mineralization [70] [67]. Categorize samples as "High" or "Low" osteogenic potential based on quantified results.
- Model Training:
- Preprocessing: Resize all images to a uniform resolution (e.g., 1024x1024 pixels). Apply data augmentation techniques (e.g., rotation, flipping) to increase dataset size and robustness.
- Training: Use a pre-trained ResNet-50 model and perform transfer learning. The model's final layer is modified for binary classification ("High" vs. "Low" potential). The model is trained using the images from step 2 as input and the labels from step 3 as the ground truth.
- Prediction: For new, unlabeled samples, input the bright-field images captured at early time points (e.g., day 1) into the trained model. The model will output a prediction of the sample's osteogenic potential.
Protocol: Non-Destructive Prediction of Muscle Stem Cell Efficiency
This protocol describes a method using phase-contrast imaging and Random Forest classification to predict the differentiation efficiency of human induced pluripotent stem cells (hiPSCs) into muscle stem cells (MuSCs) [69].
Key Reagents:
- Cells: MYF5-tdTomato reporter hiPSCs [69].
- Differentiation Media: Specific media for inducing dermomyotome and subsequent MuSC differentiation [69].
- Imaging Equipment: Phase-contrast microscope.
Procedure:
- Cell Culture and Differentiation: Induce MuSC differentiation in hiPSCs according to the established protocol, which spans approximately 82 days [69].
- Image Acquisition: Between days 14 and 38 of the differentiation process, capture phase-contrast images of the cells in culture.
- Feature Extraction:
- Apply a Fast Fourier Transform (FFT) to each cell image to obtain its power spectrum.
- Perform shell integration on the power spectrum to generate a 100-dimensional, rotation-invariant feature vector. This vector captures the essential morphological characteristics of the cells during differentiation.
- Model Training and Classification:
- On day 82, use flow cytometry to quantify the final MuSC induction efficiency (e.g., MYF5+% or CDH13 positivity) for each sample [69].
- Train a Random Forest classifier using the FFT-derived feature vectors from step 3 as input and the final efficiency measurements as labels.
- Prediction: The trained model can classify new samples as having high or low MuSC induction efficiency based on images taken weeks before the protocol endpoint.
Experimental Workflow and Signaling Pathways
The following diagram illustrates the integrated experimental and computational workflow for the early prediction of stem cell differentiation potential.
Figure 1: Workflow for ML-based prediction of differentiation potential. The process begins with stem cell culture and differentiation induction. Early morphological data is captured via non-destructive imaging and processed through feature extraction (either hand-crafted like FFT or learned via deep learning). A machine learning model uses these features to predict the final differentiation outcome, which is later validated by destructive endpoint assays [69] [67].
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Reagents and Tools for Differentiation and Analysis
| Item Name | Function/Application | Example Use Case |
|---|---|---|
| Osteogenic Induction Medium | Drives MSC commitment to osteoblastic lineage; contains inducters like dexamethasone and ascorbate-2-phosphate [70] [71]. | In vitro differentiation of MSCs into bone-forming cells for bone tissue engineering [70] [67]. |
| Adipogenic Induction Medium | Promotes MSC differentiation into adipocytes; typically includes inducters that stimulate lipid accumulation [71]. | Studying fat cell development, metabolic diseases, and for in vitro model systems [28]. |
| Chondrogenic Induction Medium | Induces MSC differentiation into chondrocytes; often includes TGF-β3 and BMP-6 [70]. | Cartilage regeneration studies and modeling chondrogenic disorders [70]. |
| MYF5-tdTomato Reporter hiPSCs | Genetically engineered hiPSCs where expression of the myogenic factor MYF5 is linked to a fluorescent tag [69]. | Tracking and quantifying muscle stem cell differentiation efficiency via fluorescence (e.g., flow cytometry) [69]. |
| CDH13 / CD105 / CD73 / CD90 Antibodies | Cell surface markers used for identifying and characterizing MSCs and specific differentiated populations like MuSCs [69] [28]. | Flow cytometry analysis to confirm cell identity and purity before and after differentiation [69] [28]. |
| Alizarin Red S / Alkaline Phosphatase (ALP) Kit | Histochemical stains for detecting calcium deposits (Alizarin Red) and ALP activity, markers of osteogenic differentiation [70] [67]. | Endpoint validation of successful osteogenesis in cell cultures [67]. |
| ACAN, FABP4, Col11a1 Primers | Gene-specific primers for quantitative RT-PCR analysis of chondrogenesis (ACAN), adipogenesis (FABP4), and osteogenesis (Col11a1) [70]. | Molecular validation of differentiation at the transcriptome level [70]. |
The fusion of quantitative morphology and machine learning represents a paradigm shift in stem cell research and manufacturing. The protocols outlined herein demonstrate that it is feasible to non-invasively predict the differentiation fate of stem cells with high accuracy, significantly earlier than traditional methods allow. This capability is crucial for enhancing the reproducibility and robustness of differentiation protocols, optimizing biomaterial design, and ultimately ensuring the quality and efficacy of stem cell-based therapies. As these computational tools continue to evolve and become more integrated into standard laboratory practice, they will undoubtedly accelerate the translation of regenerative medicine from the bench to the bedside.
The regeneration of complex musculoskele-tal tissues often requires the recapitulation of the native microenvironment, which provides not only biochemical but also physical cues, including mechanical forces and endogenous bioelectric fields [72]. Piezoelectric biomaterials, such as poly-L-lactic acid (PLLA), have emerged as a promising class of "smart" scaffolds that can dynamically convert physiological mechanical loads into localized electrical stimulation without external power sources [72] [73]. This inherent property allows them to mimic the natural electrophysiological microenvironment of tissues like bone and cartilage, which themselves exhibit piezoelectricity—the ability to generate electrical charge in response to mechanical deformation [74] [73]. Within the context of stem cell differentiation research for osteogenic, chondrogenic, and adipogenic lineages, these materials provide a powerful tool to direct cell fate. This Application Note provides a structured overview of key quantitative findings, detailed experimental protocols, and essential research tools for leveraging piezoelectric materials to enhance the differentiation of mesenchymal stem cells (MSCs).
Key Experimental Data and Findings
The following tables summarize quantitative data from pivotal studies on piezoelectric materials for osteogenic and chondrogenic differentiation of MSCs.
Table 1: Performance of Piezoelectric Scaffolds in MSC Differentiation
| Material Composition | Piezoelectric Coefficient (d33) | Cell Type | Key Differentiation Outcomes | Reference |
|---|---|---|---|---|
| Poled ZnO-PCL (10 wt% ZnO) | 0.21 ± 0.05 pC/N | Human MSCs | Under dynamic compression: Chondrogenic differentiation (higher collagen type II, GAG, Sox-9) in unpoled group; Osteogenic differentiation (higher collagen type I, VEGF-A) in poled group. | [74] |
| PCL/DPC (Bi1/2Na1/2TiO3) | Not Specified | Pre-osteoblasts | 46.3% improvement in cell proliferation rate compared to hydroxyapatite (HA) control scaffolds. | [75] |
| PCL/DPC + PCL/CPC (Hybrid) | Not Specified | Pre-osteoblasts | 7.4% improvement in osteogenic differentiation compared to PCL/HA control scaffolds. | [75] |
| Corona-poled PLLA | Not Specified | Rabbit BMSCs | Superior chondrogenic and osteogenic differentiation on piezoelectric PLLA vs. non-piezoelectric PDLLA under mechanical stimulation. | [76] |
Table 2: Summary of Stimulation Parameters and Mechanistic Insights
| Stimulation Type | Parameters | Cell Type | Mechanistic Insights | Reference |
|---|---|---|---|---|
| Piezoelectric (Quartz substrat) | Ultrasound-induced; ISATA: 1.87–14.31 mW/cm²; 5 min | BMMSC, Primary Chondrocytes | Accelerated cell migration & rearrangement via PKCζ activation; disruption of primary cilia orientation. | [77] |
| Dynamic Compression | Physiological loading regime | Human MSCs on ZnO-PCL | Piezoelectric output under load directed lineage specification: poled scaffolds favored osteogenesis, unpoled favored chondrogenesis. | [74] |
| Direct ES (C-Pace EM) | 1 Hz, 20 ms pulse, 4.6 V/cm | Fibroblasts | Activated ion channels (Piezo1), induced Ca2+ influx, increased chromatin accessibility, and promoted cell proliferation/migration. | [78] |
| Tensile Stress (in vivo) | Mandibular advancement model | Condylar Cartilage Stem/Progenitor Cells (CSPCs) | Promoted chondrogenesis via the Piezo1-Ca2+-Prkca pathway. | [79] |
Detailed Experimental Protocols
Protocol: Chondrogenic/Osteogenic Differentiation of MSCs on Piezoelectric ZnO-PCL Scaffolds Under Dynamic Compression
This protocol is adapted from the study on biodegradable zinc oxide composite scaffolds [74].
I. Scaffold Preparation and Poling
- Fabrication: Prepare a 10 wt% zinc oxide (ZnO) nanoparticle and polycaprolactone (PCL) composite. Electrospin or 3D-print the mixture to create a 3D fibrous scaffold.
- Corona Poling: To enhance piezoelectric output, subject one group of scaffolds to a high-voltage corona poling process (e.g., 10-15 kV for 30-60 minutes at elevated temperature). Leave another group unpoled for comparison.
- Characterization: Measure the piezoelectric coefficient (d33) using a piezometer. For the described scaffold, a d33 of 0.21 pC/N is expected for the poled group [74].
II. Cell Seeding and Pre-culture
- Cell Source: Expand human bone marrow-derived mesenchymal stem cells (hBMSCs) in standard growth medium (e.g., DMEM with 10% FBS and 1% penicillin/streptomycin).
- Seeding: Sterilize scaffolds (UV light or ethanol) and seed at a density of 50,000 - 100,000 cells per scaffold in a low-volume well to facilitate cell attachment.
- Pre-culture: Culture the cell-scaffold constructs for 24-48 hours to allow for full cell adhesion before applying mechanical stimulation.
III. Dynamic Compression and Differentiation
- Bioreactor Setup: Transfer the constructs to a dynamic compression bioreactor. Use a PCL-only scaffold as a control.
- Culture Medium: Use a basal differentiation medium without supplemental inductive factors (e.g., without TGF-β3 for chondrogenesis or dexamethasone for osteogenesis) to isolate the effect of piezoelectric stimulation.
- Stimulation Regime: Apply a physiological dynamic compression regime. The exact parameters (frequency, strain, duration) should be optimized for your system. A typical example is 1 Hz, 5-10% strain, for 1-2 hours per day over 14-28 days.
- Maintenance: Change the culture medium every 2-3 days. Maintain control constructs under the same conditions but without mechanical loading.
IV. Outcome Analysis
- Chondrogenic Analysis (typically stronger in unpoled ZnO-PCL):
- Biochemical: Quantify sulfated glycosaminoglycan (GAG) content using a DMMB assay. Normalize to total DNA.
- Genetic: Analyze expression of chondrogenic genes (SOX9, COL2A1, ACAN) via qRT-PCR.
- Histological: Perform Safranin-O or Alcian Blue staining on sectioned constructs to visualize proteoglycans.
- Osteogenic Analysis (typically stronger in poled ZnO-PCL):
- Biochemical: Quantify calcium deposition and mineralization using Alizarin Red S staining and extraction.
- Genetic: Analyze expression of osteogenic genes (Runx2, Osteocalcin, COL1A1) via qRT-PCR.
- Protein: Immunofluorescence staining for Collagen Type I and VEGF-A.
Protocol: Piezoelectric Stimulation of Cell Migration and Rearrangement via PKCζ
This protocol is adapted from research using ultrasound to stimulate quartz substrates [77].
I. Stimulation Chamber Setup
- Substrata: Use an AT-cut quartz coverslip (for piezoelectric stimulation) and a standard glass coverslip (for ultrasound-only control). Sterilize and coat with appropriate adhesion proteins.
- Cell Seeding: Seed bone marrow-derived MSCs (BMMSCs) or primary chondrocytes on both substrata until 80-90% confluent.
- Inhibitor Treatment (Optional): To probe mechanism, pre-treat a set of cells with 5 μM of a specific PKCζ inhibitor (e.g., ZIP pseudosubstrate peptide) for 40 minutes prior to stimulation.
II. Ultrasound and Piezoelectric Stimulation
- Apparatus: Set up a laboratory-developed ultrasonic stimulation chamber that holds the quartz/glass coverslip.
- Stimulation Parameters: Apply ultrasonic stimulation at a frequency and intensity that induces deformation in the quartz. Reported parameters are: continuous wave, ISATA of 1.87–14.31 mW/cm², for 5 minutes [77].
- Post-Stimulation: After stimulation, return cells to the standard incubator for the desired outcome period (e.g., 16-24 hours for migration assays).
III. Outcome Analysis
- Wound Healing/Migration Assay: Create a scratch wound in a confluent monolayer before stimulation. Image the wound immediately after scratching and after 16-24 hours. Quantify the percentage of wound closure.
- Cell Polarity and Rearrangement:
- Fix and immunofluorescently label cells for acetylated alpha-tubulin (cytoskeleton) and ARL13B (primary cilia).
- Use confocal microscopy to analyze the orientation and position of the primary cilium relative to the nucleus and the direction of cell migration.
- Mechanistic Validation: Analyze protein expression or activity of PKCζ via Western blot or activity assays to confirm its role.
Signaling Pathway Visualizations
The following diagrams, generated using Graphviz DOT language, illustrate key signaling pathways implicated in piezoelectric-mediated stem cell differentiation.
Diagram Title: Piezoelectric Signal Transduction in Stem Cells
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Materials and Reagents for Piezoelectric Stem Cell Research
| Item/Category | Specific Examples | Function/Application in Research |
|---|---|---|
| Piezoelectric Polymers | Poly-L-lactic acid (PLLA), Polyvinylidene fluoride (PVDF) | Flexible, biocompatible scaffold base material that generates electrical charge under mechanical deformation. [72] [76] |
| Piezoelectric Ceramics | Zinc Oxide (ZnO), Barium Titanate (BaTiO3) | High-piezoelectric-coefficient fillers for composite scaffolds to enhance electrical output. [74] [75] [73] |
| Scaffold Fabrication | Electrospinning, 3D Bioprinting | Techniques to create fibrous or porous 3D architectures that mimic the extracellular matrix. [75] |
| Poling Instrument | Corona Poling Setup | Applies a high electric field to align dipole moments within the polymer, enhancing its piezoelectric properties. [74] |
| Mechanical Bioreactors | Dynamic Compression Bioreactors | Applies controlled, physiological mechanical loads to cell-scaffold constructs to activate piezoelectricity. [74] |
| Piezoelectric Coefficient Tester | Piezometer (d33 meter) | Quantifies the piezoelectric performance (pC/N) of fabricated scaffolds. [74] |
| Key Inhibitors/Agonists | PKCζ Inhibitor (ZIP), Ca²⁺ Chelator (BAPTA-AM), Piezo1 Agonist (Yoda1) | Pharmacological tools to dissect molecular mechanisms of piezoelectric signaling. [77] [79] |
| Cell Lineage Markers | Antibodies for COL2A1, SOX9, Runx2, Osteocalcin; Alizarin Red S, Safranin-O | Used to detect and quantify osteogenic and chondrogenic differentiation outcomes. [74] [80] |
Piezoelectric biomaterials represent a paradigm shift in tissue engineering, moving from static scaffolds to dynamic, bioactive systems that actively participate in regenerative processes. The data and protocols outlined herein demonstrate that materials like PLLA and ZnO-composites can significantly enhance the osteogenic and chondrogenic differentiation of MSCs by harnessing physiologically relevant mechanical energy. The key mechanistic players, including ion channel activation, calcium influx, and downstream effectors like PKCζ, provide specific targets for optimizing future therapeutic strategies. By integrating these smart materials with standardized experimental workflows, researchers can advance the development of more effective and clinically translatable regenerative therapies for bone and cartilage repair.
Navigating Challenges: Enhancing Efficiency, Standardization, and Clinical Scalability
The therapeutic promise of mesenchymal stromal cells (MSCs) in regenerative medicine is significantly challenged by inherent donor and source variability, which profoundly impacts their differentiation potential and clinical efficacy. MSCs, once considered a homogeneous population, are now recognized as a heterogeneous group of cells whose biological properties are influenced by a complex interplay of donor characteristics including age, health status, and tissue origin [81]. This application note synthesizes current research findings to provide a structured framework for understanding and addressing these variability factors, with specific focus on osteogenic, chondrogenic, and adipogenic differentiation lineages within the broader context of stem cell differentiation research. By presenting standardized protocols and analytical approaches, we aim to equip researchers and drug development professionals with methodologies to account for and leverage this biological diversity in both experimental and therapeutic applications.
Quantitative Analysis of Donor and Source Variability
Impact of Donor Age on Differentiation Potential
Table 1: Age-Related Variations in MSC Differentiation Potential
| Age Group | Species | Tissue Source | Proliferation Capacity | Osteogenic Potential | Chondrogenic Potential | Adipogenic Potential | Reference |
|---|---|---|---|---|---|---|---|
| Fetal | Bovine | Adipose | High (30+ population doublings) | Not specified | Not specified | High | [81] |
| Calf (6-11 months) | Bovine | Adipose | High (30+ population doublings) | Not specified | Not specified | Not specified | [81] |
| Young Adult (<30 years) | Human | Adipose | Stable | Not specified | Not specified | Not specified | [82] |
| Old Adult (>50 years) | Human | Adipose | Stable | Not specified | Not specified | Not specified | [82] |
| Postmenopausal (60-81 years) | Human | Adipose | Not specified | Relatively high | Not specified | Relatively lowered | [83] |
| Newborn (0 days) | Equine | Bone Marrow | Not specified | High | High (proteoglycan content) | Not specified | [84] |
| Geriatric (≥22 years) | Equine | Bone Marrow | Not specified | Low | Low (proteoglycan content) | Not specified | [84] |
Impact of Donor Health Status and Tissue Source
Table 2: Health Status and Tissue Source Effects on MSC Properties
| Variability Factor | Specific Category | Key Findings on Differentiation Potential | Additional Characteristics | Reference |
|---|---|---|---|---|
| Health Status | Type 2 Diabetes | Greater chondrogenic potential; Lower adipogenic potential; Comparable osteogenic potential | Enhanced pro-angiogenic potential; Functional for autologous ATMPs | [85] |
| Health Status | Healthy Donors | Standard chondrogenic, adipogenic, and osteogenic potential | Baseline pro-angiogenic potential | [85] |
| Tissue Source | Bone Marrow (Equine) | Higher chondrogenic performance than AT-MSCs; Declines with age | Alkaline phosphatase activity higher than AT-MSCs | [84] |
| Tissue Source | Adipose Tissue (Equine) | Minimal chondrogenic performance; Osteogenesis affected later by age than BM-MSCs | Calcium deposition affected later by donor age | [84] |
| Breed | Holstein Friesian (Bovine) | Higher adipogenic potential (fetal and adult) | High proliferation capacity (fetal and calf) | [81] |
| Breed | Belgian Blue (Bovine) | Better osteogenic differentiation potential | Lower percentage of CD34+ cells (calf) | [81] |
| Cell Subtype (Human OA) | CD146+ sorted cells | Highest osteogenic performance (calcium deposition) | From microfragmented adipose tissue | [80] |
| Cell Subtype (Human OA) | CD271+ sorted cells | Greatest chondrogenic performance (proteoglycan formation) | From microfragmented adipose tissue | [80] |
Experimental Protocols for Assessing Differentiation Potential
Standardized Tri-Lineage Differentiation Protocol
Adipogenic Differentiation:
- Induction Medium: Complete growth medium (CGM) supplemented with 1 μM dexamethasone, 5 μg/mL insulin, 100 μM indomethacin, and 500 μM isobutylmethylxanthine [83].
- Culture Conditions: Seed cells at 5×10⁴ cells/well in 6-well plates, grow to 90-100% confluency in CGM (1-3 days), then culture in adipogenic differentiation medium (ADM) for 14 days [83].
- Analysis: Assess lipid droplet accumulation via Oil Red O staining after 14 days of induction [83].
Osteogenic Differentiation:
- Induction Medium: CGM supplemented with 50 μM ascorbic acid, 0.1 μM dexamethasone, and 10 mM β-glycerolphosphate [83].
- Culture Conditions: Seed cells at 5×10⁴ cells/well in 6-well plates, grow to 90-100% confluency in CGM (1-3 days), then culture in osteogenic differentiation medium (ODM) for 14 days [83].
- Analysis: Visualize calcium accumulation via Alizarin Red S staining after 14 days of induction [83].
Chondrogenic Differentiation:
- Induction Medium: Dulbecco's Modified Eagle media, high glucose without glutamine, supplemented with 1% FBS, 100 U/mL penicillin, 100 μg/mL streptomycin, 4 mM L-glutamine, 6.25 μg/mL insulin, 10 ng/mL transforming growth factor-β3, and 50 nM ascorbate-2-phosphate [83].
- Culture Conditions: Culture cells in chondrogenic induction media for 3-4 weeks as pellet cultures [83].
- Analysis: Fix, embed in paraffin, section, and stain with Safranin O or Alcian Blue to characterize proteoglycan content [83].
Donor Variability Assessment Workflow
Diagram 1: Experimental workflow for assessing donor variability impacts on MSC differentiation potential. This integrated approach enables systematic correlation of donor characteristics with functional differentiation outcomes.
3D Culture Differentiation Protocol
Alginate Hydrogel Chondrogenesis:
- Encapsulation: Suspend MSCs in 1.2% alginate solution at 10×10⁶ cells/mL [86].
- Cross-linking: Extrude alginate-cell mixture into 102 mM CaCl₂ solution to form hydrogel constructs [86].
- Culture: Maintain in chondrogenic induction medium for 28 days with medium changes every 2-3 days [86].
Gelatin Microribbon Hydrogel Osteogenesis/Adipogenesis:
- Seeding: Seed MSCs onto sterile gelatin µRB scaffolds at density of 1×10⁶ cells/scaffold [86].
- Culture: Maintain in appropriate osteogenic or adipogenic induction medium for 14-21 days [86].
- Analysis: Assess differentiation via tissue-specific staining and qPCR analysis [86].
Signaling Pathways in Donor-Mediated Differentiation
Diagram 2: Signaling pathways and molecular mechanisms through which donor factors influence MSC differentiation potential. Donor characteristics modulate surface marker expression, which in turn affects key signaling pathways and gene expression patterns, ultimately determining functional differentiation outcomes.
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Reagents for MSC Differentiation Studies
| Reagent/Category | Specific Examples | Function/Application | Considerations |
|---|---|---|---|
| Isolation Enzymes | Liberase, Collagenase I, Dispase | Tissue dissociation and MSC isolation | Concentration and incubation time vary by tissue source; Liberase used at 1 mg/mL for adipose tissue [81] |
| Culture Media | LG-DMEM, αMEM, DMEM/Ham's F-12 | Baseline cell culture and expansion | Supplement with FBS (10-30%) or human platelet lysate (5%) [81] [85] |
| Adipogenic Inducers | Dexamethasone, Insulin, Indomethacin, IBMX | Induction of adipogenic differentiation | Standard cocktail: 1 μM dexamethasone, 5 μg/mL insulin, 100 μM indomethacin, 500 μM IBMX [83] |
| Osteogenic Inducers | Ascorbic acid, Dexamethasone, β-glycerolphosphate | Induction of osteogenic differentiation | Standard cocktail: 50 μM ascorbic acid, 0.1 μM dexamethasone, 10 mM β-glycerolphosphate [83] |
| Chondrogenic Inducers | TGF-β3, Insulin, Ascorbate-2-phosphate | Induction of chondrogenic differentiation | Key component: 10 ng/mL TGF-β3 in pellet or 3D culture [83] |
| Differentiation Stains | Oil Red O, Alizarin Red S, Safranin O, Alcian Blue | Visualization of differentiated phenotypes | Oil Red O (lipids), Alizarin Red S (calcium), Safranin O/Alcian Blue (proteoglycans) [83] |
| Flow Cytometry Antibodies | CD34, CD73, CD90, CD105, CD146, CD271 | MSC phenotyping and population characterization | CD34 associated with proliferation; CD146 with osteogenesis; CD271 with chondrogenesis [81] [80] |
| 3D Culture Systems | Alginate hydrogels, Gelatin microribbons | Enhanced differentiation modeling | Donor variability effects differ between 2D and 3D systems [86] |
Application Notes for Research and Clinical Translation
Donor Selection Criteria
When designing studies or therapeutic approaches requiring MSCs, careful consideration of donor characteristics is essential:
- Age Considerations: For chondrogenic and osteogenic applications, younger donors (particularly neonatal sources) demonstrate superior differentiation performance [84]. However, adipose-derived stromal cells from older donors maintain angiogenic potential, suggesting lineage-specific age effects [82].
- Health Status: Diabetic donors provide functional MSCs with enhanced chondrogenic and pro-angiogenic potential, supporting autologous applications for diabetic patients [85].
- Tissue Source Selection: Bone marrow-derived MSCs exhibit superior chondrogenic potential compared to adipose-derived sources, though both are affected by donor age [84]. The tissue source should align with the target differentiation lineage.
Standardization Approaches
To address inherent donor variability in research and clinical applications:
- Superlot Generation: Create pooled donor cell populations from age-clustered donors to minimize individual variability while maintaining biological relevance [83].
- Cell Sorting Strategies: Implement magnetic-activated cell sorting (MACS) for specific subpopulations (CD146+ for osteogenesis, CD271+ for chondrogenesis) to enhance differentiation efficiency [80].
- 3D Culture Systems: Utilize biomaterial-based 3D culture platforms, as standard 2D models cannot reliably predict MSC differentiation capacity in more physiologically relevant environments [86].
Clinical Translation Protocol
MFAT Processing for Osteoarthritis Treatment:
- Tissue Harvest: Obtain subcutaneous abdominal adipose tissue via lipoaspiration under local analgesia [80].
- Microfragmentation: Process tissue using closed-system microfragmentation to generate MFAT without enzymatic digestion [80].
- Characterization: Assess stem cell subpopulations (CD34+, CD146+, CD271+) via flow cytometry to predict therapeutic potential [80].
- Application: Administer MFAT intra-articularly, with consideration for enriching specific subpopulations based on individual patient needs and disease characteristics [80].
The systematic investigation of donor and source variability factors provides both challenges and opportunities in MSC-based research and therapies. Rather than attempting to eliminate biological variability, researchers and clinicians should develop stratification protocols that align donor characteristics with specific therapeutic applications. The integration of standardized differentiation protocols, appropriate biomaterial systems, and subpopulation enrichment strategies will enhance reproducibility and efficacy in both experimental and clinical settings. As the field advances, personalized approaches that account for donor age, health status, and tissue source will be essential for unlocking the full therapeutic potential of MSC-based regenerative therapies.
The stromal vascular fraction (SVF) of tissues, such as lipoaspirate, represents a highly heterogeneous cellular source, containing subpopulations of adipose-derived stem cells (ASCs) and other progenitors with varying differentiation potentials [87]. This inherent heterogeneity presents a significant challenge for both basic science and translational, cell-based applications, as the presence of non-stem cell types can confound experimental results and therapeutic outcomes. For research focused on the osteogenic, chondrogenic, and adipogenic lineages, the isolation of a well-defined, high-potency subpopulation is a critical first step. Efficient and reliable cell sorting methods are therefore essential tools to isolate cellular subpopulations based on specific biochemical markers, enabling researchers to study differentiation mechanisms in a more controlled context [87] [44].
Comparative Analysis of Cell Sorting Technologies
The two predominant technologies for isolating cell subpopulations are Fluorescence-Activated Cell Sorting (FACS) and Magnetic-Activated Cell Sorting (MACS). The choice between them depends on the experimental requirements for purity, yield, throughput, and multi-parameter capability.
Table 1: Quantitative Comparison of FACS vs. MACS Performance
| Performance Metric | FACS | MACS |
|---|---|---|
| Cell Loss | ~70% [87] | 7-9% [87] |
| Processing Time (Single, Low-Proportion Sample) | Baseline (Slower) | 4-6 times faster than FACS [87] |
| Processing Time (Multiple Samples) | Sequential processing | Always faster overall due to parallel processing [87] |
| Multi-Marker Capability | Excellent | Not easily compatible [87] |
| Purity/Accuracy | Gold standard; high accuracy across all cell proportions [87] | Requires optimization for accuracy, especially at high target cell proportions (>~25%) [87] |
| Cell Viability | >83% [87] | >83% [87] |
Key Surface Markers for Isolating Stem Cell Subpopulations
Isolation strategies frequently rely on surface markers and intracellular enzyme activity to identify and select for stem cell subpopulations. The table below summarizes key markers used in the field.
Table 2: Key Markers and Reagents for Isolating Stem Cell Subpopulations
| Marker/Reagent | Type | Function and Relevance |
|---|---|---|
| ALPL (Alkaline Phosphatase) | Surface Marker | A membrane-bound protein involved in early matrix mineralization during osteogenesis; useful for identifying osteogenically-primed subpopulations [87]. |
| CD34 | Surface Marker | A common stem cell marker; used in combination with other markers (e.g., CD31, CD45) to define ASC populations [87]. |
| ALDH1 (Aldehyde Dehydrogenase 1) | Intracellular Enzyme | High activity is a functional marker of stem cells in various tissues; isolated cells show self-renewal, multipotency, and drug-resistance [88]. |
| CD133 | Surface Marker | An important marker present on neural stem cells and CSCs from glioblastomas, breast, prostate, and colon cancers; however, its specificity can be context-dependent [88]. |
| Osteogenic Supplements | Culture Reagent | Typically includes β-glycerophosphate, ascorbate-2-phosphate, and dexamethasone to prime cells towards the osteogenic lineage [87] [44]. |
| Anti-ALPL-APC | Antibody | Fluorescently-conjugated antibody (e.g., clone W8B2) used for labeling ALPL-expressing cells for FACS isolation [87]. |
| Microbeads (e.g., Anti-APC Microbeads) | Reagent | Magnetic beads conjugated to antibodies for indirect magnetic labeling of cells for MACS separation [87]. |
Detailed Experimental Protocols
Protocol 1: Fluorescence-Activated Cell Sorting (FACS) for ALPL+ Cells
This protocol details the isolation of ALPL+ cells from a heterogeneous mixture of osteogenically-primed SVF cells using FACS [87].
Cell Preparation and Osteogenic Priming:
- Thaw and expand passage 0 (P0) human SVF cells in expansion medium (e.g., DMEM-F12 + 10% FBS + growth factors) until >90% confluent in T182 flasks.
- Prime cells by switching to osteogenic medium (e.g., DMEM-high glucose + 10% FBS + 10 mM β-glycerophosphate + 150 µM ascorbate-2-phosphate + 10 nM dexamethasone) for four days to upregulate ALPL expression.
Cell Harvest and Labeling:
- Detach primed SVF cells and ALPL− control cells (e.g., A375 melanoma cells) using Accutase. Filter cells through a 40 µm cell strainer.
- Determine viability and cell count using trypan blue exclusion.
- Create an 8 x 10⁶ cell mixture at the desired ALPL+:ALPL− ratio (e.g., 1:3, 1:1, 3:1).
- Pellet cells and resuspend in 110 µL of a 1:11 dilution of anti-ALPL-APC antibody stock solution. Incubate for 10 minutes at 4°C.
- Keep cells on ice until sorting.
FACS Sorting:
- Use a flow cytometer (e.g., BD Influx) fitted with a 100 µm nozzle and appropriate lasers (e.g., 633 nm for APC).
- Establish FSC/SSC gates and laser gains using labeled and unlabeled control samples.
- Filter the cell mixture through a 40 µm strainer immediately before sorting.
- Sort cells into ALPL+ and ALPL− fractions, collecting into tubes containing cold base medium to preserve viability.
- Keep sorted cells on ice. Count cells and determine output fractions and viability.
Protocol 2: Magnetic-Activated Cell Sorting (MACS) for ALPL+ Cells
This protocol describes the isolation of ALPL+ cells using MACS, which offers higher yields and faster processing for some applications [87].
Cell Preparation and Priming: Perform Steps 1 and 2 from the FACS protocol (Section 4.1) to obtain a single-cell suspension of osteogenically-primed SVF cells.
Magnetic Labeling:
- Pellet the harvested and counted cells.
- Resuspend the cell pellet in a working solution of MACS Microbeads conjugated to the appropriate antibody (e.g., anti-ALPL or anti-APC for an indirect labeling approach). The manufacturer's recommended concentration may require optimization; higher concentrations may be necessary for accurate separation when the target population is abundant [87].
- Incubate for 15 minutes in the refrigerator (2-8°C).
Magnetic Separation:
- Place the MACS column in the magnetic field of the separator.
- Prepare the column by rinsing with appropriate buffer (e.g., autoMACS rinsing solution).
- Apply the cell suspension to the column.
- Collect the flow-through containing the unlabeled, ALPL− cells.
- Remove the column from the magnetic separator and elute the magnetically labeled ALPL+ cells by applying buffer and firmly pushing the plunger.
- Count the cells in both fractions and determine viability.
Protocol 3: In vitro Osteogenic Differentiation
This standard protocol is used to validate the osteogenic potential of the isolated subpopulation [44].
- Seed the isolated cells at an appropriate density (e.g., 2 x 10⁴ cells/cm²) in culture well plates.
- Culture the cells in osteogenic differentiation medium (e.g., DMEM-high glucose + 10% FBS + 10 mM β-glycerophosphate + 150 µM ascorbate-2-phosphate + 10 nM dexamethasone).
- Maintain cultures for 2-4 weeks, changing the differentiation medium every 2-3 days.
- Assess osteogenic differentiation by staining for calcium deposits with Alizarin Red S after 14-21 days.
Signaling Pathways in Mesenchymal Stem Cell Differentiation
The molecular pathways governing lineage commitment are crucial for understanding stem cell potency. The Mitogen-activated protein kinase (MAPK) pathway is a key regulator.
Integrated Workflow for Subpopulation Isolation and Characterization
The following diagram outlines a complete experimental workflow from the initial heterogeneous population to the functional characterization of the isolated subpopulation.
The directed differentiation of mesenchymal stem cells (MSCs) into osteogenic, chondrogenic, and adipogenic lineages is paramount for advancing regenerative medicine strategies for bone, cartilage, and adipose tissue repair. The inherent plasticity of MSCs, while therapeutic, presents a significant challenge: ensuring precise lineage commitment within complex biological environments. This application note details how the strategic functionalization of biomaterial interfaces—through biochemical, biophysical, and structural modifications—can mimic native stem cell niches to guide specific lineage fate. We provide a comprehensive overview of key biomaterial properties, detailed experimental protocols for assessing differentiation, visualizations of critical signaling pathways, and a curated list of essential research reagents to facilitate the development of advanced regenerative therapies.
Mesenchymal stem cells (MSCs) are multipotent stromal cells capable of self-renewal and differentiation into several mesodermal lineages, including osteoblasts, chondrocytes, and adipocytes [28]. The commitment of MSCs to a specific lineage is governed by a complex interplay of intrinsic genetic programs and extrinsic cues from the extracellular matrix (ECM) [89]. Biomaterial scaffolds, when intelligently functionalized, can replicate these ECM signals to direct cell fate. The core challenge lies in optimizing the scaffold's interface to present a defined set of signals that override default differentiation pathways and robustly steer MSCs toward a desired lineage. This document outlines the key parameters for such optimization and provides standardized protocols for validation.
Biomaterial Properties and Their Influence on Lineage Commitment
The selection and modification of biomaterials are critical, as their inherent properties directly influence cellular behavior. The table below summarizes the primary biomaterial classes and their key characteristics relevant to trilineage differentiation.
Table 1: Biomaterial Classes for Directing MSC Lineage Fate
| Material Class | Examples | Key Characteristics | Primary Lineage Influence |
|---|---|---|---|
| Natural Polymers | Collagen, Chitosan, Alginate, Gelatin, Hyaluronan, Silk Fibroin [89] | High biocompatibility, inherent bioactivity, often contain cell-adhesion motifs, enzymatically degradable. | Osteogenic (Collagen I), Chondrogenic (Hyaluronan, Collagen II) |
| Synthetic Biodegradable Polymers | Poly(L-lactic acid) (PLLA), Poly(glycolic acid) (PGA), Polycaprolactone (PCL), Poly(lactic-co-glycolic acid) (PLGA) [89] | Tunable mechanical properties, controllable degradation rates, consistent batch-to-batch quality. | Osteogenic (stiff substrates), Adipogenic (soft substrates) |
| Conductive Polymers | Polypyrrole, Polyaniline, Polythiophene [89] | Conduct electrical impulses, enhance neurite outgrowth (in neural applications), can be used for electrical stimulation. | Osteogenic (with electrical stimulation) |
The functionalization of these materials involves modifying their surfaces or bulk properties to introduce specific signals. Key functionalization strategies include:
- Biochemical Functionalization: Covalent grafting or physical adsorption of bioactive molecules such as specific peptides (e.g., RGD for adhesion), growth factors (e.g., BMP-2 for osteogenesis, TGF-β3 for chondrogenesis), or small molecules.
- Biophysical Functionalization: Engineering surface topography (e.g., nanogrooves, pits), substrate stiffness (elastic modulus), and ligand density to provide mechanotransductive cues.
- Structural Functionalization: Designing 3D scaffold architectures with controlled porosity, pore size, and interconnectivity to influence nutrient diffusion, cell-cell contact, and tissue ingrowth.
Quantitative Assessment of Trilineage Differentiation
To validate the efficacy of a functionalized biomaterial, robust quantitative assessment of MSC differentiation is essential. The following table outlines key molecular markers and functional assays for each lineage.
Table 2: Key Markers and Assays for Assessing MSC Differentiation
| Lineage | Key Genetic Markers | Functional Assays (In Vitro) | Critical Signaling Pathways |
|---|---|---|---|
| Osteogenesis | Col11a1 (early marker) [70], Runt-related transcription factor 2 (RUNX2), Alkaline Phosphatase (ALP) | Alizarin Red S staining for calcium deposition, ALP activity assay, von Kossa staining for mineralization [28] [70] | BMP/Smad, Wnt/β-catenin, ERK signaling [89] |
| Chondrogenesis | ACAN (Aggrecan) [70], Collagen type II (COL2A1), SOX9 | Alcian Blue or Safranin O staining for proteoglycan content, immunohistochemistry for COL2A1, pellet culture system [28] [70] | TGF-β/Smad, ERK/Stat6, MERTK signaling [89] |
| Adipogenesis | FABP4 (Fatty Acid-Binding Protein 4) [70], Peroxisome Proliferator-Activated Receptor Gamma (PPARγ), Lipoprotein Lipase (LPL) | Oil Red O staining for lipid droplet formation, glycerol-3-phosphate dehydrogenase (GPDH) activity assay [28] [70] | PPARγ signaling, cAMP-mediated pathways |
Experimental Protocols
Protocol 4.1: Biomaterial Functionalization via Peptide Conjugation
This protocol describes a method for covalently grafting the RGD peptide to a chitosan scaffold to enhance cell adhesion.
- Scaffold Preparation: Create porous chitosan scaffolds via freeze-drying. Sterilize by immersion in 70% ethanol for 2 hours, followed by extensive washing with sterile phosphate-buffered saline (PBS).
- Surface Activation: Incubate scaffolds in a 50 mM MES buffer (pH 5.5) containing 50 mg/mL 1-Ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC) and 25 mg/mL N-Hydroxysuccinimide (NHS) for 30 minutes at room temperature with gentle agitation to activate carboxyl groups.
- Peptide Conjugation: Rinse activated scaffolds with cold MES buffer. Transfer to a solution of 1 mM RGD peptide (sequence: GRGDS) in MES buffer and incubate for 4 hours at 4°C.
- Quenching and Washing: Terminate the reaction by incubating scaffolds in 1M ethanolamine solution (pH 8.5) for 1 hour. Wash thoroughly with PBS and store in PBS at 4°C until cell seeding.
Protocol 4.2: Real-Time Quantitative PCR (rt-qPCR) for Differentiation Marker Analysis
This protocol, adapted from validated kits, quantifies the expression of key differentiation markers [70].
- Cell Seeding and Differentiation: Seed human Adipose-Derived Stem Cells (ADSCs) at passage 3-4 onto functionalized biomaterials at a density of 50,000 cells/cm². Culture in standard growth medium until 80% confluent.
- Induction: Replace growth medium with specific differentiation induction media:
- Osteogenic: DMEM supplemented with 100 nM dexamethasone, 10 mM β-glycerolphosphate, and 0.05 mM ascorbate-2-phosphate [70].
- Adipogenic: DMEM supplemented with 1 μM dexamethasone, 0.5 mM isobutylmethylxanthine, 10 μg/mL insulin, and 200 μM indomethacin.
- Chondrogenic: High-glucose DMEM supplemented with 10 ng/mL TGF-β3, 100 nM dexamethasone, 0.1 mM ascorbate-2-phosphate, and 1x ITS+ premix [70].
- Maintain cultures for 14-21 days, changing the induction medium every 3-4 days.
- RNA Extraction: Lyse cells directly on the scaffold using TRIzol reagent. Isolate total RNA according to the manufacturer's instructions. Assess RNA purity and concentration using a spectrophotometer.
- cDNA Synthesis: Synthesize cDNA from 1 μg of total RNA using a reverse transcription kit with oligo(dT) primers.
- rt-qPCR Amplification: Prepare reactions using SYBR Green master mix, gene-specific primers (e.g., ACAN, FABP4, Col11a1), and cDNA template. Run in triplicate on a real-time PCR system. Use the following cycling conditions: 95°C for 10 min; 40 cycles of 95°C for 15 sec and 60°C for 1 min.
- Data Analysis: Calculate relative gene expression using the 2−ΔΔCT method. Normalize target gene expression to a housekeeping gene (e.g., GAPDH) and report as fold-change relative to undifferentiated control cells cultured on the same scaffold material.
Signaling Pathway Visualizations
Diagram 1: Key signaling pathways directing MSC trilineage differentiation. External stimuli (e.g., growth factors) activate receptor-mediated pathways, leading to the upregulation of key transcription factors (colored nodes) that drive the expression of lineage-specific marker genes.
Diagram 2: A generalized workflow for developing and validating a functionalized biomaterial for directed stem cell differentiation, from initial fabrication to final data analysis.
The Scientist's Toolkit: Research Reagent Solutions
The following table catalogs essential materials and reagents required for experiments in biomaterial-driven stem cell differentiation.
Table 3: Essential Research Reagents for Biomaterial and Differentiation Studies
| Reagent/Material | Function/Application | Example Specification / Note |
|---|---|---|
| Adipose-Derived Stem Cells (ADSCs) | Primary cell source for differentiation studies; multipotent. | Isolate from stromal vascular fraction (SVF); confirm expression of CD105, CD73, CD90, and lack of CD45, CD34 [28] [70]. |
| Chitosan | Natural polymer scaffold; biocompatible, modifiable. | >75% deacetylation; use for creating porous 3D scaffolds via freeze-drying. |
| Polycaprolactone (PCL) | Synthetic polymer scaffold; offers tunable mechanical properties. | Mn ~80,000; suitable for electrospinning or 3D printing. |
| RGD Peptide | Functionalization agent; promotes integrin-mediated cell adhesion. | Sequence: GRGDS; >95% purity; conjugate to scaffold via EDC/NHS chemistry. |
| Recombinant Human TGF-β3 | Soluble induction factor for chondrogenic differentiation. | Use at 10 ng/mL in chondrogenic induction medium [70]. |
| Dexamethasone | Synthetic glucocorticoid; component of all three lineage induction media. | Use at 100 nM for osteogenesis, 1 μM for adipogenesis, 100 nM for chondrogenesis [70]. |
| TRIzol Reagent | Monophasic solution for the isolation of high-quality total RNA from cells on scaffolds. | - |
| rt-qPCR Kit (SYBR Green) | For quantitative analysis of differentiation marker gene expression. | Validate primers for efficiency; use 2−ΔΔCT method for analysis [70]. |
| Alizarin Red S | Histochemical stain for detecting calcium deposits in osteogenic cultures. | Quantify by elution and spectrophotometry. |
| Oil Red O | Histochemical stain for detecting neutral lipids and lipid droplets in adipogenic cultures. | Quantify by elution and spectrophotometry. |
| Alcian Blue | Histochemical stain for detecting sulfated proteoglycans in chondrogenic cultures. | - |
The application of machine learning (ML) to predict stem cell differentiation into osteogenic, chondrogenic, and adipogenic lineages represents a frontier in regenerative medicine and drug development. These computational models promise to accelerate the development of cell-based therapies by predicting cell fate from high-dimensional data, such as transcriptomics and cellular morphology [90] [28]. However, two significant challenges impede their reliable application in biological research: data standardization and model interpretability. Without robust standardization, biological data from different sources and formats introduce noise that compromises model performance [91] [92]. Furthermore, the "black box" nature of complex models, like deep learning, obstructs biological insight, making it difficult for researchers to trust and learn from the model's predictions [93]. This application note provides detailed protocols and frameworks to address these dual challenges, enabling the development of more accurate, reliable, and biologically insightful prediction models.
Data Standardization Protocols for Stem Cell Research
The Critical Need for Standardization
In stem cell research, data is generated from diverse sources—including single-cell RNA sequencing, microscopy images, and flow cytometry—each with inherent variations in format, scale, and structure. Inconsistent data directly impacts model performance; for example, a study predicting mesenchymal stem cell (MSC) differentiation achieved 94.7% accuracy using standardized morphological data, a feat impossible with raw, unprocessed inputs [94]. Standardization transforms raw, heterogeneous data into a consistent, analysis-ready format, forming the foundation for any predictive modeling endeavor [92].
A Step-by-Step Standardization Framework
The following multi-stage protocol ensures data is uniformly structured for optimal model training.
Step 1: Define Data Standards and a Common Data Model (CDM)
- Action: Establish a project-specific data dictionary defining the standard format for all data elements.
- Protocol:
- Biological Entities: Define standard naming conventions for genes (e.g., official HGNC symbols), proteins, and cell markers (e.g., CD90, CD105, CD73 for MSCs [28]).
- Experimental Data: Standardize units for gene expression counts (e.g., Transcripts Per Million), differentiation scores, and image metadata.
- Metadata: Document sample preparation protocols, donor information, and passage number using controlled vocabularies.
- Output: A centralized, accessible data dictionary documenting all standards.
Step 2: Profile and Audit Existing Data
- Action: Systematically scan all data sources to identify inconsistencies.
- Protocol:
- Use data profiling tools or custom scripts to analyze the distribution of values, missing data rates, and format variations.
- For transcriptome data, identify variations in gene symbol nomenclature (e.g., "Oct4" vs. "Pou5f1").
- For image data, audit variations in resolution, file format, and staining intensities.
- Output: A data quality report quantifying inconsistencies and missing values.
Step 3: Cleanse and Prepare Data
- Action: Correct identifiable errors and prepare data for transformation.
- Protocol:
- Remove technical duplicates from sequencing datasets.
- Correct obvious typos in categorical data (e.g., "ostegenic" to "osteogenic").
- In imaging data, remove out-of-focus images or artifacts.
- Output: A cleansed dataset ready for transformation.
Step 4: Apply Standardization Rules and Transform Data
- Action: Convert all data to the standards defined in Step 1.
- Protocol:
- Numeric Data: Apply unit conversions and scale data (e.g., Z-score normalization for gene expression values).
- Textual Data: Convert all text to a consistent case (e.g., uppercase), expand abbreviations and align on common terms (e.g., "hMSC" for human mesenchymal stem cells) [92].
- Date/Time Formats: Convert all timestamps to ISO 8601 format (e.g.,
YYYY-MM-DD) for tracking differentiation time courses [95]. - Image Data: Standardize dimensions, resolution, and color channels. Apply normalization (e.g., pixel values to a 0-1 range).
Step 5: Validate and Review Standardized Data
- Action: Quality control check to ensure transformation accuracy.
- Protocol:
- Run automated validation scripts to check adherence to the CDM.
- Manually review a data sample against source records.
- For quantitative data, use positive and negative control samples to confirm expected outcomes.
- Output: A validation report and a finalized, analysis-ready dataset.
Standardization Workflow
The following diagram illustrates the complete data standardization pathway from raw biological data to a model-ready dataset.
Interpretability Frameworks for Biological Insight
The Need for Explainability in Stem Cell Models
While complex models like convolutional neural networks (CNNs) and graph neural networks offer high predictive accuracy, their decisions are often opaque. Explainable AI (XAI) methods are crucial for translating model predictions into testable biological hypotheses, moving beyond a "black box" [93]. For instance, an interpretable deep learning model applied to single-cell transcriptomic data of aging discovered a novel ribosomal gene subnetwork and an inflammatory response pathway, insights that were missed by standard models [93].
Protocol for Implementing Model Interpretability
This protocol integrates XAI into the model development lifecycle for stem cell prediction models.
Step 1: Integrate Biological Networks as Prior Knowledge
- Action: Incorporate existing biological knowledge to ground the model and its explanations in reality.
- Protocol:
- Network Selection: Obtain a protein-protein interaction (PPI) network or a signaling pathway database (e.g., from KEGG, Reactome).
- Data Integration: Overlay your standardized data (e.g., gene expression) onto this network. Each gene is a node, and interactions are edges [93].
- Outcome: The model learns from data within the context of known biological relationships, making subsequent explanations more meaningful.
Step 2: Select and Train a Predictive Model with Explainability in Mind
- Action: Choose a model architecture compatible with specific XAI techniques.
- Protocol:
- For Image Data (Morphology): Use a pre-trained CNN (e.g., ResNet50, VGG19) for tasks like classifying osteogenic vs. adipogenic differentiation from microscopy images [94].
- For Transcriptomic/Network Data: Use a Graph Convolutional Network (GCN) to learn from the PPI-integrated data [93].
- Training: Train the model on standardized data to predict differentiation outcomes or chronological age.
Step 3: Apply Explainable AI (XAI) Techniques
- Action: Extract post-hoc explanations from the trained model.
- Protocol:
- For CNNs (Image Data): Use Gradient-weighted Class Activation Mapping (Grad-CAM) to generate heatmaps highlighting image regions (cell structures) most influential in the prediction.
- For GNNs (Network Data): Use methods like PGExplainer to identify the most predictive edges (interactions) and nodes (genes) within the biological network [93].
- Output: Visual explanations (heatmaps, highlighted sub-networks) and ranked lists of important features.
Step 4: Validate Biological Interpretations Experimentally
- Action: Bridge the computational-biological gap by testing model-derived insights.
- Protocol:
- Hypothesis Generation: Formulate a hypothesis based on XAI output (e.g., "Gene X is critical for osteogenic differentiation").
- Experimental Validation: Use knockdown (e.g., siRNA) or overexpression of highlighted genes in stem cells and assess the impact on differentiation using standard assays (e.g., qPCR for lineage markers, staining).
- Iterate: Use validation results to refine the model.
Interpretable Analysis Workflow
The diagram below outlines the integrated workflow for building and interpreting predictive models using biological networks and XAI.
Experimental Validation & Performance Metrics
Quantitative Performance of Standardized Models
Implementing the described protocols for data standardization and interpretability directly enhances model performance. The table below summarizes quantitative results from key studies in the field.
Table 1: Performance Metrics of Standardized and Interpretable ML Models in Stem Cell Research
| Study Focus | Model Architecture | Key Standardization Steps | Interpretability Method | Performance Metric | Result |
|---|---|---|---|---|---|
| Predicting hMSC differentiation (osteogenic vs. adipogenic) from morphology [94] | ResNet50 (CNN) | Image normalization, sizing, data augmentation | Gradient-based attention maps | Classification Accuracy | 95.7% (Binary), 94.7% (Multi-class) |
| VGG19 (CNN) | Image normalization, sizing, data augmentation | Gradient-based attention maps | Classification Accuracy | ~95% (Binary & Multi-class) | |
| Building aging clocks from single-cell transcriptomics [93] | Multi-view Graph Representation Learning (MGRL) | Gene symbol standardization, PPI network integration, meta-cell formation | PGExplainer | Mean Absolute Error (MAE) in predicting chronological age | 8.50 years (outperformed ElasticNet and Random Forest benchmarks) |
| Classifying pluripotent stem cells [90] | Convolutional Neural Network (CNN) | Standardized brightfield imaging, morphological feature extraction | Model confidence scores | Classification Accuracy | >85% |
Experimental Protocol: Validating an Interpretable Differentiation Predictor
This protocol provides a detailed methodology for replicating a morphology-based deep learning experiment, as referenced in [94].
Aim: To predict osteogenic and adipogenic differentiation of human Mesenchymal Stem Cells (hMSCs) using live-cell imaging and a convolutional neural network (CNN).
Materials:
- Cell Line: Human bone marrow-derived MSCs (e.g., from Lonza or PromoCell).
- Culture Medium: Mesenchymal Stem Cell Basal Medium (MSCBM) supplemented with growth factors.
- Differentiation Media: Commercial osteogenic and adipogenic induction media.
- Imaging Equipment: Phase-contrast or brightfield microscope with live-cell imaging capabilities (e.g., an incubator-housed microscope).
- Computational Resources: Workstation with GPU (e.g., NVIDIA Tesla series) and deep learning frameworks (e.g., TensorFlow, PyTorch).
Method:
- Cell Culture and Differentiation:
- Culture hMSCs in standard growth medium until ~70% confluency.
- Split cells and seed into imaging plates.
- For differentiation groups, replace growth medium with osteogenic or adipogenic induction media. Maintain a control group in growth medium.
- Change the media every 2-3 days.
Live-Cell Image Acquisition:
- Begin imaging 24 hours after induction.
- Acquire images from multiple, pre-defined fields of view in each well at regular intervals (e.g., every 24 hours) for the duration of the experiment (e.g., 15 days).
- Maintain constant imaging conditions (exposure time, magnification, light intensity).
Image Data Standardization & Labeling:
- Standardize Images: Resize all images to a uniform pixel size (e.g., 224x224). Apply normalization to scale pixel values to a standard range (e.g., 0-1).
- Create Dataset: Label each image with its corresponding class (e.g., "Osteogenic-Day7", "Adipogenic-Day10", "Control-Day5"). This is crucial for supervised learning.
Model Training & Interpretation:
- Model Selection: Choose a pre-trained CNN model (e.g., ResNet50) and perform transfer learning.
- Training: Split the standardized image dataset into training, validation, and test sets. Train the model to classify the images.
- Interpretation: Apply Grad-CAM to the trained model. This will produce heatmaps superimposed on the input images, highlighting the cellular regions (e.g., lipid vacuoles in adipocytes, mineralized nodules in osteocytes) that most strongly influenced the model's classification decision.
Validation:
- Correlative Analysis: At the end of the live-cell experiment, fix the cells and perform standard confirmatory assays:
- Adipogenesis: Oil Red O staining for lipid droplets.
- Osteogenesis: Alizarin Red S staining for calcium deposits.
- Correlate the staining results with the model's predictions and Grad-CAM heatmaps to verify biological accuracy.
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Research Reagents and Computational Tools for ML-Driven Stem Cell Research
| Item Name | Function/Application | Specification Notes |
|---|---|---|
| Human Mesenchymal Stem Cells (hMSCs) | Primary model system for studying osteogenic, adipogenic, and chondrogenic differentiation. | Source: Bone marrow, adipose tissue. Must be characterized for CD105+, CD73+, CD90+, CD34-, CD45-, HLA-DR- [28]. |
| Osteogenic & Adipogenic Induction Media | Induces lineage-specific differentiation for model training and validation. | Typically contains dexamethasone, ascorbate, and β-glycerophosphate (osteogenic) or insulin, indomethacin, and IBMX (adipogenic). |
| Protein-Protein Interaction (PPI) Database | Provides prior biological knowledge for graph-based models and interpretability. | Examples: STRING, BioGRID. Used to build molecular networks for analysis [93]. |
| Pre-trained CNN Models (e.g., ResNet50, VGG19) | Base architecture for transfer learning on image-based differentiation prediction. | Pre-trained on large image datasets (e.g., ImageNet), allowing for efficient adaptation to biological images [94]. |
| Graph Neural Network (GNN) Models (e.g., DeeperGCN) | Analyzes structured, network-based data like integrated transcriptomics and PPI. | Capable of learning from graph-structured data where nodes have feature attributes [93]. |
| Explainable AI (XAI) Tools (e.g., PGExplainer, Grad-CAM) | Provides post-hoc interpretations of complex model predictions. | PGExplainer for GNNs; Grad-CAM for CNNs. Critical for extracting biological insights [93] [94]. |
The directed differentiation of stem cells into osteogenic, chondrogenic, and adipogenic lineages represents a cornerstone of regenerative medicine. However, a significant translational challenge persists: conventional two-dimensional (2D) in vitro culture systems often fail to replicate the complex physiological conditions cells experience in vivo, leading to functionally immature or aberrant phenotypes [96]. The native stem cell microenvironment, or niche, is a complex three-dimensional (3D) space comprising not only biochemical cues but also essential physical and mechanical signals [97]. These mechanical elements—including extracellular matrix (ECM) topology, substrate stiffness, and fluid shear stress—are not merely passive scaffolds but active regulators of cell fate [97]. This protocol details methodologies to bridge the in vitro to in vivo gap by engineering microenvironments that faithfully mimic these native conditions, thereby promoting the maturation of functionally robust bone, cartilage, and fat cells from mesenchymal stem/stromal cells (MSCs) for research and drug development applications.
The Mechanical Microenvironment: Components and Biological Significance
The mechanical microenvironment is an integral component of the stem cell niche, working in concert with biochemical factors to direct differentiation. The table below summarizes the key mechanical factors and their specific roles in guiding MSC fate.
Table 1: Key Mechanical Factors in the Stem Cell Microenvironment and Their Influence on MSC Differentiation
| Mechanical Factor | Description | Impact on MSC Differentiation |
|---|---|---|
| Extracellular Matrix (ECM) & Topology | A 3D network of macromolecules (e.g., collagen, fibronectin); topology refers to the surface geometry/nanostructure [97]. | 3D culture better simulates the native environment, altering biological behavior. Nanotopography can enhance osteogenesis; pore size influences differentiation trajectory (e.g., smaller pores enhance osteogenesis) [97]. |
| Substrate Stiffness | The mechanical rigidity of the surface to which cells adhere [97]. | A dominant factor. Stiffer substrates (e.g., ~10-40 kPa) promote osteogenic differentiation. Softer substrates (e.g., ~0.5-2 kPa) promote adipogenic and chondrogenic differentiation [97]. |
| Shear Stress | The frictional force exerted by fluid flow over the cell surface [97]. | Simulated fluid flow in dynamic culture systems can promote the expression of specific lineage progenitors and enhance differentiation maturity [97]. |
| Cell Morphology & Geometry | The physical shape and spatial constraints imposed on a cell [97]. | Morphological changes can regulate stem cell signaling and differentiation fate. Spreading and flattening often favor osteogenesis, while a more rounded morphology favors adipogenesis and chondrogenesis [97]. |
Experimental Protocols for Engineering the Microenvironment
Protocol 1: Fabricating 3D Scaffolds with Controlled Stiffness and Topology for Osteogenic and Chondrogenic Differentiation
Principle: This protocol utilizes biomaterial scaffolds to provide a 3D mechanical milieu that directs stem cell fate. By varying the scaffold composition and architecture, one can control substrate stiffness and topological features like pore size to guide differentiation towards osteogenic or chondrogenic lineages [97].
Materials:
- Polycaprolactone (PCL): A biodegradable polymer for 3D printing scaffolds [97].
- Alginate/Graphene Oxide: Components for creating composite hydrogels with tunable stiffness [97].
- β-Tricalcium Phosphate (β-TCP): A ceramic material with ECM-like properties that promotes osteogenesis [97].
- Cell-derived ECM (dECM): ECM harvested from cultured cells to biofunctionalize scaffolds [97].
Method:
- Scaffold Fabrication:
- For osteogenic promotion, fabricate a stiff, porous scaffold using 3D printing of PCL or a composite of alginate/graphene oxide [97].
- For chondrogenic promotion, create a softer hydrogel scaffold with a larger pore size to support cell aggregation into islet-like clusters [97].
- Optional Biofunctionalization: Coat the scaffold with cell-derived ECM (e.g., osteoblast-derived ECM for bone, chondrocyte-derived ECM for cartilage) to enhance bioactivity and lineage-specific differentiation [97].
Cell Seeding and Culture:
- Seed human MSCs (hMSCs) onto the scaffold at a density of 1-5 million cells/mL.
- Maintain cells in standard growth medium for 24-48 hours to allow for attachment and proliferation.
Induction of Differentiation:
- Replace the growth medium with osteogenic or chondrogenic induction medium.
- For osteogenic induction, use medium supplemented with dexamethasone, β-glycerophosphate, and ascorbic acid.
- For chondrogenic induction, use medium supplemented with TGF-β3, dexamethasone, ascorbic acid, and ITS+ premix.
- Culture for 14-21 days, changing the medium every 2-3 days.
Protocol 2: Modulating Substrate Stiffness using Tunable Hydrogels for Adipogenic and Osteogenic Differentiation
Principle: This protocol employs hydrogels with tunable mechanical properties to investigate the dose-dependent effect of substrate stiffness on the adipogenic-osteogenic fate decision of MSCs [97].
Materials:
- Tunable Hydrogels (e.g., Alginate, Polyacrylamide): Commercially available hydrogel kits that allow for precise control of elastic modulus.
- Stiffness Modulators: Crosslinking agents (e.g., CaCl₂ for alginate) to vary hydrogel stiffness.
Method:
- Hydrogel Preparation:
- Prepare a series of hydrogels with stiffness values spanning from soft (0.5 kPa) to stiff (40 kPa), following manufacturer protocols.
- For adipogenic differentiation, use soft hydrogels (0.5-2 kPa).
- For osteogenic differentiation, use stiff hydrogels (10-40 kPa) [97].
Cell Seeding:
- Seed hMSCs at a standard density (e.g., 10,000 cells/cm²) onto the surface of the pre-formed hydrogels.
Differentiation and Analysis:
- Culture cells in a standardized differentiation medium capable of supporting both lineages, or in lineage-specific induction media.
- Incubate for 7-14 days. The substrate stiffness will biasing the differentiation outcome.
- Analysis: Fix and stain cells for lineage-specific markers: Oil Red O for adipogenic lipids (adipogenesis) and Alizarin Red S for calcium deposits (osteogenesis).
Quantitative Assessment of Differentiation Outcomes
Validating the success of microenvironment engineering requires quantitative assessment of lineage-specific markers. Transcriptomic analyses provide a powerful tool for this purpose. The following table summarizes key genes whose expression is indicative of successful differentiation into osteogenic, adipogenic, and chondrogenic lineages.
Table 2: Key Gene Expression Markers for Monitoring MSC Differentiation
| Gene Symbol | Gene Name | Function / Significance | Expression in Differentiation |
|---|---|---|---|
| Runx2 | Runt-related transcription factor 2 | Master regulator of osteoblast differentiation; activates osteoblast-specific genes [98]. | Osteogenic |
| Ocn | Osteocalcin | Non-collagenous protein found in bone; marker of mature osteoblasts [98]. | Osteogenic |
| PPARγ | Peroxisome proliferator-activated receptor gamma | Master regulator of adipogenesis; controls adipocyte-specific genes and inhibits osteogenesis [98]. | Adipogenic |
| Lamc1 | Laminin subunit gamma 1 | Encodes a component of laminins in the ECM; promotes osteogenic differentiation and inhibits adipogenic differentiation [98]. | Osteogenic Adipogenic |
| Col4a1 | Collagen type IV alpha 1 chain | A major component of the basement membrane; associated with ECM-receptor interactions during differentiation [98]. | Co-expressed |
| Hexb | Beta-hexosaminidase subunit beta | Involved in glycolipid biosynthesis; upregulated during osteogenic differentiation [98]. | Osteogenic |
Signaling Pathways Governing Mechanotransduction and Fate Decisions
Stem cells perceive mechanical cues from their environment through mechanosensors and convert them into biochemical signals, a process known as mechanotransduction. The pathway below illustrates the key molecular events triggered by a stiff substrate, leading to osteogenic differentiation.
Diagram 1: Mechanotransduction from Stiff Substrate to Osteogenesis.
The balance between osteogenesis and adipogenesis is tightly regulated by competing signaling pathways and transcription factors. Key pathways like Wnt/β-catenin promote osteogenesis, while PPARγ activation is the central driver of adipogenesis.
Diagram 2: Transcriptional Regulation of Osteogenic vs. Adipogenic Fate.
The Scientist's Toolkit: Essential Research Reagent Solutions
Successfully implementing these protocols requires a suite of reliable reagents and tools. The following table details essential solutions for research in this field.
Table 3: Essential Research Reagent Solutions for Microenvironment-Mimicking Studies
| Research Reagent / Tool | Function / Application | Example Use Case |
|---|---|---|
| Tunable Hydrogels (Alginate, PA) | To create substrates with defined and physiologically relevant stiffness for 2D and 3D culture. | Investigating the effect of substrate stiffness (0.5-40 kPa) on the adipogenic-osteogenic fate switch [97]. |
| 3D Bioprinting / Electrospinning Systems | To fabricate scaffolds with controlled architecture, porosity, and topology. | Producing PCL or PLA scaffolds with specific pore sizes to enhance osteogenic differentiation or support chondrogenic aggregation [97]. |
| CRISPR-Cas9 System | For precise genome editing to introduce or correct disease-associated mutations in stem cells. | Creating genetically defined iPSC lines for disease modeling or knocking out genes like LAMC1 to validate its functional role in differentiation [99] [100]. |
| Cell-derived ECM (dECM) | A biologically active coating that mimics the native niche to enhance attachment and direct differentiation. | Coating synthetic PCL scaffolds with osteoblast-derived dECM to potentiate the osteogenic differentiation of hMSCs [97]. |
| Small Molecule Inhibitors/Activators | To chemically manipulate key signaling pathways involved in differentiation. | Using a GSK-3β inhibitor to activate Wnt/β-catenin signaling and promote osteogenesis over adipogenesis [98]. |
| scRNA-Seq Kits | To characterize cellular heterogeneity and transcriptomic changes at a single-cell resolution during differentiation. | Profiling the distinct subpopulations within an organoid or 3D culture to assess differentiation efficiency and identity [99] [98]. |
The transition of stem cell research from laboratory discovery to clinical therapy hinges on the development of robust, scalable manufacturing processes that consistently produce high-quality, functional differentiated cells. For mesenchymal stem cells (MSCs) directed toward osteogenic, chondrogenic, and adipogenic lineages—central to regenerative strategies for bone, cartilage, and adipose tissue disorders—this manufacturing challenge is particularly complex [28] [33]. These lineages share a common mesenchymal origin and exist in a delicate developmental balance, meaning manufacturing protocols must not only efficiently drive differentiation but also rigorously control lineage specificity and purity [98] [33]. The ultimate goal is to establish standardized, clinically compliant processes that can generate the billions of cells required for widespread therapeutic application, a feat that demands the integration of advanced bioprocessing technologies with stringent quality control systems [101].
This document outlines key strategies and provides detailed protocols for the scalable manufacturing and quality assessment of clinical-grade osteocytes, chondrocytes, and adipocytes derived from stem cells, framed within the context of current regulatory and industrial landscapes.
Key Challenges in Scaling Differentiated Cell Manufacturing
Scaling the production of differentiated stem cells for clinical use presents a unique set of challenges that move beyond conventional laboratory practice.
- Process Variability and Raw Materials: The quality and characteristics of starting materials, such as patient or donor-derived cells, exhibit inherent variability. Furthermore, raw materials, media, and reagents can introduce batch-to-batch variations, leading to unpredictable yields and critical quality attributes (CQAs) [102].
- Lineage Crosstalk and Contamination: The differentiation pathways for osteogenic, chondrogenic, and adipogenic lineages are interconnected and often antagonistic. For instance, peroxisome proliferator-activated receptor γ (PPARγ), a master regulator of adipogenesis, can inhibit osteogenesis [28]. In chondrogenic micromass cultures, the use of dexamethasone has been shown to inadvertently induce adipogenic differentiation in human synovium-derived stem cells [33]. This crosstalk necessitates precise control to ensure final product purity.
- Tumorigenicity Risk: The use of pluripotent stem cells (PSCs), including induced PSCs (iPSCs), as a starting source introduces the critical risk of residual undifferentiated cells, which can form tumors in vivo [103]. Establishing robust purification and detection methods for these cells is a non-negotiable safety requirement.
- Scalability and Automation: Many research-grade differentiation protocols are manual, labor-intensive, and conducted in 2D flask systems, which are ill-suited for producing the vast cell numbers needed for clinical trials and commercial therapy. Transitioning to 3D bioreactor systems is essential but can alter cell behavior, affecting transfection efficiency, differentiation efficacy, and final product quality [101] [102].
Quality by Design (QbD) and Critical Quality Attributes (CQAs)
A Quality by Design (QbD) framework is fundamental to developing a robust manufacturing process. This involves defining a Quality Target Product Profile (QTPP) and identifying CQAs that are linked to critical process parameters (CPPs) through structured Design of Experiment (DoE) studies [101] [102].
Table 1: Critical Quality Attributes (CQAs) for Differentiated Mesenchymal Lineages
| Lineage | Morphology | Key Molecular Markers | Functional Assays | Purity/Safety |
|---|---|---|---|---|
| Osteogenic | Mineralized matrix nodules, cuboidal shape | Up: RUNX2, Osteocalcin, ALP, COL1A1 [98] | Calcium deposition (Alizarin Red S), ALP activity [98] | ≤ 0.001% residual undifferentiated PSCs [103] |
| Chondrogenic | Round/elliptical, lacunae formation in 3D pellets | Up: SOX9, Aggrecan (ACAN), COL2A1 [28] [33] | Sulfated glycosaminoglycan (sGAG) content (Alcian Blue/Safranin O) [28] | Absence of hypertrophic markers (e.g., COL10A1) [98] |
| Adipogenic | Intracellular lipid vacuoles (signet-ring) | Up: PPARγ, C/EBPα, FABP4 [28] [33] | Lipid accumulation (Oil Red O staining) [28] | >95% cells FABP4+ by flow cytometry |
For all lineages derived from pluripotent sources, a universal CQA is the absence of tumorigenic cells. Metabolic selection methods (e.g., using glucose- and glutamine-depleted media to selectively eliminate undifferentiated human PSCs while sparing differentiated cardiomyocytes) or small molecules like PluriSIn have proven effective for large-scale, non-invasive purification [103].
Scalable Bioprocessing Strategies
Moving from static 2D culture to scalable, controlled bioreactor systems is pivotal for clinical and commercial manufacturing.
- Upstream Process Development: Micro and mini bioreactor systems (e.g., Ambr systems) are invaluable for high-throughput process optimization with minimal resource use. They enable DoE to define CPPs like pH, dissolved oxygen, and feeding schedules for differentiation [102]. These processes can be scaled to larger single-use bioreactors (e.g., from 2L to 2000L systems), which are ideal for cGMP manufacturing [102]. For example, a scalable workflow for iPSC expansion has been successfully transferred from an Ambr 250 system to 2L Univessel Glass Bioreactors, yielding billions of high-quality cells for differentiation into lineages like cardiomyocytes [102].
- Cell-Specific Considerations:
- Adherent Cells (MSCs, iPSCs): Microcarrier-based bioreactor cultures allow for the high-density expansion of adherent cells in 3D suspension, dramatically increasing yield over 2D flasks [102].
- 3D Differentiation (Chondrogenesis): Pellet or aggregate-based chondrogenic differentiation is inherently suited to bioreactor systems, which improve nutrient and oxygen mass transfer compared to static pellet cultures, promoting more uniform and higher-quality cartilage tissue formation [28].
- Downstream Processing: Scalable technologies like single-use tangential flow filtration (TFF) and counterflow centrifugation (e.g., kSep systems) are essential for the low-shear clarification, concentration, and formulation of the final cell product [102].
Detailed Experimental Protocols
Protocol 1: Scalable Osteogenic Differentiation of BMSCs in a Bioreactor
This protocol describes the osteogenic induction of bone marrow-derived MSCs (BMSCs) in a controlled, scalable bioreactor system.
Materials:
- Cells: Clinical-grade human BMSCs (passage 3-5).
- Basal Medium: DMEM, high glucose, GlutaMAX.
- Osteogenic Inducers: Ascorbic acid (50 µg/mL), β-glycerophosphate (10 mM), Dexamethasone (100 nM) [98].
- Bioreactor: Stirred-tank single-use bioreactor (e.g., Sartorius Biostat STR).
- Analytical Reagents: Alizarin Red S solution, ALP staining kit, RNA extraction kit.
Method:
- Cell Expansion: Expand BMSCs on microcarriers in the bioreactor. Maintain culture in growth medium (basal medium + 10% FBS) at 37°C, 5% CO₂, with controlled agitation (e.g., 60 rpm) to prevent aggregation.
- Induction: Upon reaching 80% confluence, switch to osteogenic induction medium. Continuously monitor and control CPPs: pH (7.4), DO (40%).
- Medium Exchange: Perform 50% medium exchanges every 2-3 days.
- Harvesting: After 14-21 days, dissociate cells from microcarriers using a validated enzyme solution (e.g., Accutase).
- Quality Control:
- Alizarin Red Staining: Fix a cell sample and stain with 2% Alizarin Red S (pH 4.2) for 20 min to visualize calcium deposits.
- ALP Activity: Quantify using a colorimetric assay (e.g., pNPP substrate) and normalize to total protein.
- qPCR Analysis: Isolve RNA and perform qPCR for RUNX2 and Osteocalcin [98]. Expression should be upregulated >20-fold compared to undifferentiated controls.
Protocol 2: Metabolic Purification of iPSC-Derived Cells
This protocol utilizes glucose- and glutamine-depleted medium to selectively eliminate residual undifferentiated iPSCs from a differentiated population [103].
Materials:
- Cells: Crude differentiation culture (e.g., iPSC-derived osteogenic progenitors).
- Purification Medium: DMEM without glucose or glutamine, supplemented with galactose, fatty acids, and lactate.
- Control Medium: Standard culture medium with glucose and glutamine.
Method:
- Preparation: At the end of the differentiation protocol, dissociate cells into a single-cell suspension.
- Metabolic Selection: Seed cells at a defined density in purification medium. Culture for 3-5 days.
- Control Culture: Seed a parallel cell sample in control medium.
- Assessment: After the selection period, compare cell viability and recovery between purification and control cultures. Non-CMs and residual iPSCs, which are highly dependent on glucose and glutamine, will undergo cell death, enriching the desired differentiated population [103].
- Validation: Use droplet digital PCR (ddPCR) to quantify the presence of pluripotency markers (e.g., OCT4) pre- and post-purification to confirm the depletion of undifferentiated iPSCs [103].
Signaling Pathways Governing Trilineage Differentiation
The fate of MSCs toward osteogenic, chondrogenic, or adipogenic lineages is governed by a complex interplay of signaling pathways. Key pathways include TGF-β/BMP, Wnt, and Hedgehog, which often have antagonistic effects on different lineages.
Diagram 1: Signaling pathway crosstalk in MSC differentiation shows TGF-β/BMP promoting both chondrogenesis (via SOX9) and osteogenesis (via RUNX2). Wnt and Hedgehog signaling strongly promote osteogenesis while inhibiting adipogenesis (via PPARγ). An antagonistic relationship exists between the osteogenic and adipogenic lineages, where RUNX2 and PPARγ mutually inhibit each other [28] [98] [33].
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Reagents for Clinical-Grade Differentiation
| Reagent/Category | Example Product(s) | Function in Differentiation | Clinical-Grade Consideration |
|---|---|---|---|
| Cell Source | REPROCELL StemRNA Clinical Seed iPSCs [104] | GMP-compliant, integration-free master cell bank provides a consistent, scalable starting material. | Drug Master File (DMF) submitted to FDA streamlines IND applications. |
| Basal Media | Xeno-free DMEM/F-12, Essential 8 | Provides fundamental nutrients; xeno-free formulation eliminates pathogen and immunogen risk. | Must be USP/EP tested and supplied with comprehensive CoA. |
| Growth Factors | Recombinant human TGF-β3, BMP-2, BMP-7 [33] | Key signaling molecules to direct chondrogenic (TGF-β3) and osteogenic (BMP-2) differentiation. | High purity (>95%), animal-free, recombinant origin is mandatory. |
| Small Molecules | Dexamethasone, Ascorbic Acid, IBMX | Synergize with growth factors; Dexamethasone is a common component in adipogenic and osteogenic cocktails. | Must be sourced as GMP Active Pharmaceutical Ingredients (APIs). |
| Culture Surfaces | Microcarriers (e.g., Cytodex), GMP-grade Matrigel | Provide a scalable 3D surface for adherent cell expansion and differentiation in bioreactors. | Full traceability and validation for absence of animal pathogens. |
| Purification Aids | PluriSIn, Metabolic Selection Media [103] | Selectively eliminates residual undifferentiated pluripotent stem cells, enhancing product safety. | Process must be validated for removal efficiency and lack of impact on differentiated cells. |
The successful manufacturing of clinical-grade differentiated cells for the osteogenic, chondrogenic, and adipogenic lineages is a multidisciplinary endeavor. It requires a deep understanding of developmental biology, translated into controlled, scalable bioprocesses under a rigorous QbD framework. By leveraging scalable bioreactor systems, implementing lineage-specific differentiation protocols with metabolic purification, and adhering to evolving regulatory standards, researchers can overcome the current challenges of variability, scalability, and safety. The continued advancement of these strategies, as evidenced by recent clinical trial progress and regulatory approvals [104], is essential for turning the promise of stem cell-based regenerative medicine into tangible therapies for patients.
Comparative Efficacy and Clinical Fidelity: Validating Sources, Potency, and Therapeutic Potential
The selection of an optimal mesenchymal stem cell (MSC) source is a critical determinant of success in regenerative medicine and tissue engineering. This application note provides a systematic comparison of the differentiation potential of four prominent MSC types: Bone Marrow-derived MSCs (BMSCs), Adipose-derived Stem Cells (ADSCs), Synovium-derived Stem Cells (SDSCs), and Infrapatellar Fat Pad-derived Stem Cells (IFPSCs). We synthesize quantitative data on their osteogenic, chondrogenic, and adipogenic capacities, present standardized protocols for their isolation and differentiation, and analyze key signaling pathways governing lineage commitment. Within the context of a broader thesis on stem cell differentiation, this resource aims to equip researchers and drug development professionals with the experimental frameworks and analytical tools necessary for informed cell source selection in musculoskeletal research and therapeutic development.
Mesenchymal stem cells (MSCs) represent a cornerstone of regenerative medicine due to their multipotent differentiation capacity, self-renewal potential, and relative ease of isolation from various tissues [105]. The International Society for Cellular Therapy (ISCT) has established minimal criteria for defining MSCs, including plastic-adherence, expression of specific surface markers (CD73, CD90, CD105), lack of hematopoietic markers (CD45, CD34, CD14, CD11b, CD19, HLA-DR), and tri-lineage differentiation potential into osteoblasts, chondrocytes, and adipocytes [106] [85] [107]. However, MSCs derived from different tissue sources exhibit significant functional heterogeneity in their proliferation rates, differentiation bias, and response to biochemical and biophysical cues [106] [105] [107].
This application note directly addresses the critical need for a cross-comparative analysis of four clinically relevant MSC sources. BMSCs were the first discovered and most extensively characterized population [106]. ADSCs have emerged as an attractive alternative due to their abundant availability and less invasive harvesting procedure [106] [108]. SDSCs have gained attention for their exceptional chondrogenic potential, making them promising for cartilage repair [109] [110]. IFPSCs, residing within the knee joint, represent a specialized adipose population with reported progenitor cell characteristics. Understanding the unique biological properties and differentiation biases of these cell types is fundamental to designing effective cell-based therapies for bone, cartilage, and adipose tissue regeneration.
Comparative Analysis of Differentiation Potential
A comprehensive analysis of peer-reviewed studies reveals distinct differentiation profiles for each MSC source. The data below summarize their relative performance in osteogenic, chondrogenic, and adipogenic lineages.
Table 1: Comparative Differentiation Potential of MSC Sources
| MSC Source | Osteogenic Potential | Chondrogenic Potential | Adipogenic Potential | Key Characteristics |
|---|---|---|---|---|
| BMSCs | High [107] [106] | High [107] | Moderate [107] | Considered the "gold standard"; osteogenic potential may decline with donor age [106]. |
| ADSCs | Moderate (inferior to BMSCs in vitro) [106] [107] | Moderate [107] | High [107] | High cell yield from lipoaspirates, faster proliferation, less discomfort during harvesting [106]. |
| SDSCs | Not fully quantified | Very High [109] | Not fully quantified | Stable chondrogenic phenotype; expressed higher levels of collagen type II and aggrecan than chondrocytes allocated for ACI [109]. |
| IFPSCs | Information not available in search results | Information not available in search results | Information not available in search results | A specialized adipose depot; often grouped with ADSCs in broader analyses. |
Table 2: Impact of Donor and Culture Conditions on MSC Potency
| Factor | Impact on Differentiation Potential |
|---|---|
| Donor Age | Conflicting reports; some studies show reduced osteogenesis in BMSCs from aged donors [106], while others show age-independent osteogenesis in ADSCs [106] [105]. |
| Disease State (e.g., Diabetes) | AT-MSCs from diabetic donors showed comparable osteogenic capacity but greater chondrogenic and pro-angiogenic potential compared to those from healthy donors [85]. |
| Cell Passaging | Chondrogenic markers (Collagen type II, Aggrecan) on SDSCs declined with further passaging [109]. CD34 expression on ASCs decreases after extensive passage [106]. |
| Culture Microenvironment | Decellularized ECM from fetal SDSCs rejuvenated chondrogenic potential in adult SDSCs, linked to MAPK and non-canonical Wnt signaling [110]. |
Experimental Protocols
Isolation and Expansion of MSCs
Protocol 1: Isolation of Human BMSCs [107]
- Collection: Obtain bone marrow aspirate (e.g., 10 mL from iliac crest) using a heparinized syringe to prevent clotting.
- Processing: Filter the aspirate through a 70 μm cell strainer and dilute 1:1 with culture medium (DMEM + 10% FBS + 1% Penicillin/Streptomycin).
- Centrifugation: Centrifuge at 1800 rpm for 10 minutes at room temperature (RT).
- Plating: Resuspend the cell pellet in culture medium and plate in a 75 cm² culture flask.
- Culture: Maintain at 37°C in a humidified atmosphere of 5% CO₂.
- Medium Change: After 24 hours, wash with PBS to remove non-adherent cells. Change the culture medium twice weekly.
- Expansion: Passage cells at 80-90% confluence using Trypsin/EDTA. Use cells at passages 3-5 for experiments.
Protocol 2: Isolation of Human ADSCs [107]
- Collection: Obtain subcutaneous adipose tissue.
- Washing: Wash tissue extensively with PBS containing 5% antibiotics.
- Digestion: Mince tissue and digest with 0.1% collagenase type I in PBS for 60 minutes at 37°C with agitation.
- Neutralization: Neutralize collagenase with an equal volume of culture medium (DMEM + 10% FBS).
- Centrifugation: Centrifuge at 2000 rpm for 5 minutes. The pellet contains the stromal vascular fraction (SVF).
- Plating and Expansion: Resuspend the SVF pellet in culture medium, plate in a 75 cm² flask, and expand as described for BMSCs.
Multi-Lineage Differentiation Assays
Protocol 3: Osteogenic Differentiation [107]
- Seeding: Seed cells (e.g., ASCs or BMSCs) at a standard density (e.g., 5x10³ cells/cm²) in growth medium.
- Induction: When cells reach 90-100% confluence, replace the growth medium with commercial osteogenic induction medium (e.g., StemPro Osteogenesis Differentiation Kit). This typically contains dexamethasone, ascorbate, and β-glycerophosphate.
- Maintenance: Culture for 2-4 weeks, changing the differentiation medium every 3 days.
- Analysis:
- Alizarin Red S Staining: Fix cells with 4% PFA and stain with Alizarin Red S to detect calcium deposits.
- Alkaline Phosphatase (ALP) Activity: Monitor ALP activity as an early osteogenic marker.
- Gene Expression: Analyze expression of osteogenic genes (e.g., Runx2, Osteocalcin) via RT-qPCR.
Protocol 4: Chondrogenic Differentiation [108]
- Pellet Culture: Resuspend 5x10⁵ cells in 0.5 mL of complete chondrogenic induction medium (e.g., MesenCult Chondrogenic Differentiation Medium).
- Centrifugation: Transfer the cell suspension to a 15 mL polypropylene tube and centrifuge at 300 ×g for 10 minutes to form a pellet.
- Induction: Loosen the tube caps and incubate at 37°C under 5% CO₂ for 3 days.
- Feeding: Add an additional 0.5 mL of induction medium to a final volume of 1 mL. Continue incubation for up to 28 days, changing the medium every three days.
- Analysis:
- Histology: Fix the cartilage pellet in formalin, embed in paraffin, section, and stain with Alcian Blue to detect sulfated glycosaminoglycans (GAGs).
- Gene Expression: Analyze expression of chondrogenic genes (e.g., Collagen type II, Aggrecan).
Protocol 5: Adipogenic Differentiation [107]
- Seeding: Seed cells at a high density (e.g., 2x10⁴ cells/cm²) in growth medium.
- Induction: At 100% confluence, replace the growth medium with commercial adipogenic induction medium (e.g., MesenCult Adipogenic Differentiation Medium), typically containing insulin, indomethacin, IBMX, and dexamethasone.
- Maintenance: Culture for 2-4 weeks, changing the medium every 3-4 days.
- Analysis:
- Oil Red O Staining: Fix cells with 4% PFA and stain with Oil Red O to visualize intracellular lipid droplets.
- Gene Expression: Analyze expression of adipogenic genes (e.g., PPARγ, FABP4).
Signaling Pathways Governing Differentiation
The differentiation of MSCs into osteogenic, chondrogenic, and adipogenic lineages is regulated by a complex interplay of conserved signaling pathways. Key pathways include TGF-β/BMP, Wnt, and Hedgehog signaling.
The Scientist's Toolkit: Essential Reagents and Materials
Table 3: Research Reagent Solutions for MSC Differentiation Studies
| Reagent/Category | Specific Examples | Function & Application |
|---|---|---|
| Isolation Enzymes | Collagenase Type I [108] [107], Dispase [37] | Digest extracellular matrix to liberate cells from native tissue (e.g., adipose, synovium). |
| Culture Media | αMEM [85], DMEM/F12 [108] | Basal media for cell expansion and maintenance. |
| Serum Supplements | Fetal Bovine Serum (FBS) [107], Human Platelet Lysate (hPL) [85] | Provides essential growth factors and adhesion proteins for cell proliferation. |
| Osteogenic Inducers | Dexamethasone, Ascorbate, β-Glycerophosphate [105] (e.g., StemPro Osteogenesis Kit [108]) | Cocktail to induce osteogenic differentiation and matrix mineralization. |
| Chondrogenic Inducers | TGF-β, Dexamethasone, Ascorbate (e.g., MesenCult Chondrogenic Kit [108]) | Induces chondrogenic differentiation in pellet or 3D culture. |
| Adipogenic Inducers | Insulin, IBMX, Indomethacin, Dexamethasone (e.g., MesenCult Adipogenic Kit [107]) | Cocktail to induce adipogenic differentiation and lipid droplet formation. |
| Characterization Antibodies | Anti-CD73, CD90, CD105 [85] [107]; Anti-CD34, CD45 [108] [107] | Flow cytometry analysis to confirm MSC immunophenotype per ISCT criteria. |
| Differentiation Stains | Alizarin Red S [108] [107], Alcian Blue [108], Oil Red O [108] [107] | Histochemical stains to detect calcium (osteogenesis), GAGs (chondrogenesis), and lipids (adipogenesis). |
The choice of MSC source should be dictated by the specific therapeutic or research objective. The comparative data and protocols provided herein serve as a guide for this decision-making process.
- For Bone Regeneration: BMSCs remain the gold standard due to their superior osteogenic capacity [106] [107]. However, ADSCs should be considered when a less invasive harvest and larger cell yield are priorities, potentially enhanced with osteogenic growth factors or genetic modification [106] [105].
- For Cartilage Repair: SDSCs demonstrate a clear advantage with their robust and stable chondrogenic phenotype, expressing high levels of native cartilage matrix proteins like collagen type II and aggrecan [109]. They represent a promising alternative to dedifferentiated passaged chondrocytes in Autologous Chondrocyte Implantation (ACI).
- For Disease Modeling and Angiogenesis: ADSCs, particularly from diabetic donors, show enhanced pro-angiogenic potential, making them suitable for modeling metabolic disease and developing related therapies [85].
In conclusion, while all MSC sources share core characteristics, their distinct differentiation biases and functional properties underscore the absence of a one-size-fits-all cell source. This application note provides a foundational framework for selecting the most appropriate MSC type based on empirical evidence and offers standardized protocols to ensure rigorous, reproducible research in stem cell-based regenerative medicine. Future work will focus on further elucidating the potential of IFPSCs and refining strategies to control and enhance MSC fate through modulation of the microenvironment and signaling pathways.
Within stem cell research and drug development, the rigorous validation of multilineage differentiation—specifically into osteogenic, chondrogenic, and adipogenic lineages—is a critical step. Functional assays provide direct, visual, and quantitative evidence of successful differentiation, moving beyond gene expression analysis to confirm phenotypic and metabolic changes [111]. These assays, including the detection of alkaline phosphatase (ALP) activity, mineralized nodules, lipid droplets, and glycosaminoglycan (GAG) deposition, are indispensable for characterizing stem cell fate. They are widely applied in basic research, tissue engineering, and preclinical studies for conditions like osteoporosis and osteoarthritis [112] [113]. This document provides detailed application notes and standardized protocols for these key functional assays, framed within the context of a comprehensive thesis on stem cell differentiation.
The Scientist's Toolkit: Essential Reagents and Kits
The following table summarizes key reagents and kits commonly used for functional validation of stem cell differentiation, as cited in recent literature.
Table 1: Key Research Reagent Solutions for Differentiation Assays
| Item Name | Function / Application | Examples from Literature |
|---|---|---|
| Osteogenic Differentiation Kit | Provides pre-mixed components for consistent induction of osteogenesis. | MesenCult Osteogenic Differentiation Kit (Stemcell Technologies) [114]; Cyagen Biosciences Osteogenic Differentiation Kit [113]. |
| Adipogenic Differentiation Kit | Provides pre-mixed components for consistent induction of adipogenesis. | MesenCult Adipogenic Differentiation Kit (Stemcell Technologies) [114]; Cyagen Biosciences Adipogenic Differentiation Kit [113]. |
| Chondrogenic Differentiation Kit | Provides pre-mixed components for consistent induction of chondrogenesis in pellet or micromass culture. | Cyagen Biosciences Chondrogenic Differentiation Kit [113]. |
| ALP Assay Kit | Quantifies alkaline phosphatase activity, an early marker of osteogenic differentiation. | Alkaline Phosphatase Activity Test Kit (Beyotime, China) [113]. |
| Alizarin Red S (ARS) | Stains calcium deposits and mineralized nodules in late-stage osteogenic cultures. | Component of osteogenic assay kits (e.g., Cyagen Biosciences); used in fixed-cell staining [113] [115]. |
| Oil Red O | Stains neutral lipid droplets in mature adipocytes. | Oil Red O Staining Kit (Beyotime, China) [113]. |
| Alcian Blue | Stains sulfated glycosaminoglycans (GAGs) in the cartilaginous extracellular matrix. | Alcian Blue Staining Kit (Beyotime, China) [113]. |
Osteogenic Differentiation Assays
Alkaline Phosphatase (ALP) Activity Assay
Principle: ALP is an early-stage enzyme highly expressed by committed osteoprogenitor cells. Its activity is a key indicator of ongoing osteogenic commitment [116].
Detailed Protocol:
- Cell Seeding and Induction: Seed BMSCs or other mesenchymal stem cells (MSCs) at a density of (5 \times 10^3) cells/cm² in multi-well plates. Culture until 70-80% confluent, then replace the growth medium with osteogenic induction medium (OIM). A typical OIM consists of Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 10 mM β-glycerophosphate, 50 μg/mL L-ascorbic acid, and 0.1 μmol/L dexamethasone [116] [113].
- Sample Collection: After 4-7 days of induction, wash the cells with phosphate-buffered saline (PBS) and lyse them using an appropriate lysis buffer (e.g., RIPA buffer) [113].
- Activity Measurement: Use a commercial ALP assay kit (e.g., from Beyotime or Nanjing Jiancheng Bioengineering Institute). Mix the cell lysate with the kit's working solution, incubate as per instructions, and measure the absorbance at 405-520 nm using a microplate reader [116] [113].
- Data Normalization: Normalize the ALP activity to the total protein concentration of the lysate, determined by a BCA Protein Assay Kit [113].
Quantitative Data: Table 2: Representative ALP Activity and Mineralization Data from Osteogenic Studies
| Cell Type | Treatment / Condition | ALP Activity (Relative Expression) | Mineralization (ARS, Relative Level) | Citation |
|---|---|---|---|---|
| hBMSCs | Control (Non-induced) | Baseline | Baseline | [116] |
| hBMSCs | Osteogenic Induction | Significantly Increased | Significantly Increased | [116] |
| hBMSCs | TSC22D3 Downregulation | Significant Decrease | Significant Decrease | [116] |
| hBMSCs | FTO Overexpression | Not Reported | Decreased | [113] |
| UCMSCs | ARNT2 Overexpression | Enhanced | Enhanced | [114] |
Mineralized Nodule Staining with Alizarin Red S (ARS)
Principle: In the late stages of osteogenesis, cells deposit calcium phosphate crystals, forming mineralized nodules. ARS binds to calcium ions, producing a characteristic red-orange stain [114] [115].
Detailed Protocol:
- Culture and Induction: Induce osteogenic differentiation for 14-21 days, changing the OIM every 2-3 days [114].
- Fixation: After the induction period, aspirate the medium, wash the cells gently with PBS, and fix with 4% paraformaldehyde for 30 minutes at room temperature [114].
- Staining: Remove the fixative, wash with distilled water, and add a 2% (w/v) Alizarin Red S solution (pH 4.1-4.3) to cover the cell layer. Incubate for 20-30 minutes at room temperature, protected from light [115].
- Washing and Imaging: Carefully aspirate the stain and wash extensively with distilled water to remove non-specific staining. Observe and capture images under a brightfield microscope. Orange-red deposits indicate mineralized nodules.
- Quantification (Optional): For semi-quantification, the stained nodules can be dissolved in 10% (w/v) cetylpyridinium chloride solution for 30 minutes, and the eluent's absorbance measured at 562 nm [115].
Adipogenic Differentiation Assay
Lipid Droplet Staining with Oil Red O
Principle: Upon adipogenic differentiation, MSCs accumulate triglycerides in intracellular lipid vacuoles. Oil Red O is a fat-soluble dye that stains these neutral lipids bright red [113] [114].
Detailed Protocol:
- Cell Seeding and Induction: Seed MSCs at a high density ((5 \times 10^4) cells/well in a 6-well plate) and allow them to reach 100% confluence. Replace the medium with adipogenic induction medium. This medium typically contains supplements like dexamethasone, indomethacin, and insulin [113]. Culture for 2-3 weeks, changing the medium every 3 days.
- Fixation: Wash the cells with PBS and fix with 4% paraformaldehyde for 30 minutes.
- Staining: Prepare a working Oil Red O solution by diluting a stock solution (0.5% in isopropanol) with distilled water (typically 3:2 ratio) and filter it. After fixing and washing, add the working solution to the cells and incubate for 30-60 minutes.
- Washing and Imaging: Remove the stain and wash thoroughly with distilled water. Counterstain the nuclei with hematoxylin if desired. Image under a brightfield microscope. Red-stained droplets within the cytoplasm indicate successful adipogenesis.
- Quantification (Optional): Stain can be eluted with 100% isopropanol and the absorbance measured at 510 nm [113].
Chondrogenic Differentiation Assay
Glycosaminoglycan Staining with Alcian Blue
Principle: Differentiated chondrocytes secrete a rich extracellular matrix (ECM) abundant in sulfated glycosaminoglycans (GAGs). Alcian Blue binds electrostatically to these polyanionic GAGs, producing a blue-green color [113].
Detailed Protocol:
- Pellet Culture and Induction: Chondrogenesis is often induced in a 3D pellet culture system to mimic the cellular condensation phase. Centrifuge (2 \times 10^5) to (5 \times 10^5) MSCs in a conical tube to form a pellet. Culture the pellet in a chondrogenic differentiation medium containing TGF-β3, dexamethasone, and ascorbate for 3-4 weeks [113].
- Fixation: Collect the pellets, wash in PBS, and fix in 4% paraformaldehyde for several hours.
- Paraffin Processing and Sectioning: Dehydrate the fixed pellets through a graded ethanol series, clear in xylene, embed in paraffin, and section into 5-10 μm thick slices using a microtome.
- Staining: Deparaffinize and rehydrate the sections. Stain with 1% Alcian Blue solution (in 3% acetic acid, pH 2.5) for 30 minutes.
- Washing and Counterstaining: Rinse the slides in running tap water to remove excess stain. Counterstain the nuclei with a neutral red or nuclear fast red solution.
- Imaging: Dehydrate, clear, and mount the sections. Visualize under a microscope. A blue-green ECM indicates the presence of GAGs and successful chondrogenesis.
Experimental Workflow and Key Signaling Pathways
The following diagram illustrates the integrated experimental workflow for the simultaneous induction and validation of trilineage differentiation from a single source of Mesenchymal Stem Cells (MSCs), incorporating the key assays described in this document.
The functional assays detailed herein—ALP activity, Alizarin Red S, Oil Red O, and Alcian Blue staining—form the cornerstone of phenotypic validation in stem cell differentiation research. When employed alongside molecular techniques like qPCR, they provide a robust framework for confirming lineage-specific differentiation. The standardized protocols and quantitative benchmarks offered in this document are designed to enhance experimental reproducibility and reliability, thereby supporting advancements in regenerative medicine and therapeutic development.
Within stem cell research and regenerative medicine, the rigorous molecular validation of differentiated cell phenotypes is paramount. For human Mesenchymal Stem/Stromal Cells (MSCs), which can differentiate into osteogenic, chondrogenic, and adipogenic lineages, this validation typically involves assessing lineage-specific marker expression and characterizing secreted protein profiles [28] [44]. This document provides detailed application notes and protocols for two cornerstone techniques: quantitative Reverse Transcription PCR (qRT-PCR) for analyzing key transcriptional markers of differentiation, and proteomic analysis for profiling the secretome. These methodologies are essential for confirming successful differentiation in academic research, quality control in biomanufacturing, and the development of cell-based therapeutics [104].
qRT-PCR for Lineage-Specific Marker Analysis
qRT-PCR is a highly sensitive and quantitative method for measuring the expression of genes associated with specific cell lineages. Accurate results depend on proper experimental design, including the selection of stable reference genes and validated lineage-specific markers.
Selection and Validation of Reference Genes
The use of inappropriate reference genes is a major source of error in qRT-PCR data normalization. It is critical to select genes with stable expression under your specific experimental conditions. A study on Inonotus obliquus underscores this principle, systematically evaluating 11 candidate reference genes under various culture conditions and finding the most stable gene differed depending on the treatment (e.g., VPS for varying carbon sources, RPB2 for different nitrogen sources) [117]. While this study was in a fungal model, it highlights a universal best practice: reference genes must be validated for your specific cell type and differentiation protocol. The stability of candidate genes should be evaluated using algorithms like GeNorm, NormFinder, and BestKeeper [117].
Lineage-Specific Markers for MSC Differentiation
The following table summarizes key transcription factors and marker genes used to validate the trilineage differentiation of MSCs.
Table 1: Key Marker Genes for MSC Differentiation Lineages
| Lineage | Gene Symbol | Gene Name | Primary Function/Role |
|---|---|---|---|
| Osteogenic | RUNX2 | Runt-Related Transcription Factor 2 | Master regulator of osteoblast differentiation; activates genes for bone matrix deposition [98]. |
| Osteogenic | OCN (BGLAP) | Osteocalcin | Non-collagenous protein found in bone; late-stage marker of osteoblast maturation and mineralization [98]. |
| Osteogenic | ALPL | Alkaline Phosphatase | Enzyme critical for bone mineralization; early marker of osteogenic commitment [98]. |
| Adipogenic | PPARγ | Peroxisome Proliferator-Activated Receptor Gamma | Master regulator of adipogenesis; promotes expression of adipocyte-specific genes and inhibits osteogenesis [98]. |
| Adipogenic | LPL | Lipoprotein Lipase | Enzyme involved in lipid metabolism; adipogenic marker [29]. |
| Adipogenic | FABP4 (aP2) | Fatty Acid Binding Protein 4 | Involved in intracellular fatty acid transport; adipogenic marker [29]. |
| Chondrogenic | SOX9 | SRY-Box Transcription Factor 9 | Key transcription factor regulating chondrocyte differentiation and cartilage formation [28]. |
| Chondrogenic | COL2A1 | Collagen Type II Alpha 1 Chain | Major structural component of cartilage extracellular matrix [28]. |
| Chondrogenic | ACAN | Aggrecan | Large proteoglycan essential for cartilage load-bearing capacity [28]. |
Key Signaling Pathways and Molecular Regulation
The commitment of MSCs to a specific lineage is governed by complex signaling pathways that often act in opposition. A primary regulatory axis exists between osteogenesis and adipogenesis. Activation of the ERK MAP kinase pathway is crucial for driving osteogenic differentiation, while its inhibition can push cells toward an adipogenic fate [29]. The Wnt/β-catenin and BMP/Smad pathways are also potent activators of osteoblast differentiation and simultaneously inhibit adipogenesis [98]. Conversely, the activation of PPARγ is the central event in adipogenesis and inhibits osteogenic differentiation [98].
Recent research has identified Lamc1 (Laminin subunit gamma-1) as a novel regulator. It is upregulated during osteogenic differentiation and downregulated during adipogenic differentiation. Functional studies show that knockdown of Lamc1 inhibits both osteogenic and adipogenic differentiation, highlighting its importance in the differentiation process and the interplay with the extracellular matrix (ECM) [98]. The ECM itself is a critical regulator, with components like collagen promoting osteogenesis and fibronectin potentially favoring adipogenesis [98].
Diagram 1: Signaling pathways in MSC differentiation. Key pathways promoting osteogenesis (green), adipogenesis (red), and chondrogenesis (blue) are shown. A key antagonistic relationship exists between the osteogenic master regulator RUNX2 and the adipogenic master regulator PPARγ.
Detailed qRT-PCR Protocol
Title: qRT-PCR Analysis of Differentiation Markers in Human MSCs
Objective: To extract high-quality RNA, synthesize cDNA, and quantify the expression of lineage-specific markers via qRT-PCR to validate MSC differentiation.
Materials:
- Differentiated and undifferentiated control MSCs
- RNA extraction kit (e.g., Ultrapure RNA Kit)
- Nanodrop spectrophotometer or equivalent
- High-capacity cDNA reverse transcription kit
- RT-qPCR instrument (e.g., ViiA7)
- qPCR master mix (e.g., SYBR Green)
- Validated primers for target and reference genes
Method:
- RNA Extraction:
- Lyse cells directly in culture plate wells using a guanidinium thiocyanate-based lysis buffer.
- Follow manufacturer's protocol for RNA purification, including DNase I treatment to remove genomic DNA contamination.
- Elute RNA in nuclease-free water.
RNA Quality and Quantity Assessment:
- Determine RNA concentration and purity (A260/A280 ratio ~2.0) using a Nanodrop.
- Verify RNA integrity by running an aliquot on a 1% agarose gel; sharp ribosomal RNA bands (28S and 18S) indicate good integrity.
cDNA Synthesis:
- Use 100 ng – 1 µg of total RNA for reverse transcription in a 20 µL reaction.
- Use a kit containing reverse transcriptase, random hexamers, dNTPs, and RNase inhibitor.
- Typical reaction conditions: 25°C for 10 min (primer annealing), 37-50°C for 30-120 min (elongation), 85°C for 5 min (enzyme inactivation).
Quantitative PCR:
- Prepare reaction mix per sample: 10 µL SYBR Green Master Mix, 0.4 µL each forward and reverse primer (10 µM), 8.2 µL nuclease-free water, 1 µL cDNA.
- Run samples in technical triplicates.
- Use a thermal cycling protocol similar to: 95°C for 5 min; 40 cycles of 95°C for 10 s, 60°C for 20 s, 72°C for 20 s; followed by a melt curve stage (95°C for 15 s, 60°C for 1 min, 95°C for 15 s) to check amplification specificity [117].
Data Analysis:
- Calculate primer amplification efficiency (E) using a standard curve of a serially diluted cDNA pool. E = [10^(-1/slope) - 1] x 100%. Ideal efficiency is 90-110%.
- Normalize target gene Ct values to the geometric mean of one or more validated reference genes (e.g., GAPDH, RPLPO).
- Analyze relative gene expression using the 2^(-ΔΔCt) method.
Proteomic Analysis of Secreted Factors (Secretome)
The secretome—the complete set of proteins secreted by a cell—provides critical insights into how MSCs communicate with their environment, influencing tissue repair, immunomodulation, and disease progression [118] [119]. Senescence-associated secretory phenotype (SASP) and cancer secretome analysis are key areas of application [118].
Methodological Challenges and an Improved Normalization Strategy
A major challenge in secretome analysis is accurate protein quantification for equal loading in mass spectrometry. The standard bicinchoninic acid (BCA) assay can overestimate protein concentration in concentrated culture media due to interfering substances, leading to inconsistent loading and compromised quantitative accuracy [118].
Solution: Concentration Rate-Based Normalization
- Concentrate conditioned media using ultrafiltration devices.
- Record the volume before (Vi) and after (Vf) concentration.
- Calculate the Concentration Rate (CR) as CR = Vi / Vf.
- Instead of relying solely on BCA, adjust the volume of the concentrated sample loaded for digestion based on the CR to ensure a more consistent and accurate protein amount across samples [118]. This method has been shown to improve reproducibility and reliability in secretome profiling.
Detailed Protocol for Secretome Analysis
Title: Data-Independent Acquisition (DIA) Mass Spectrometry for Secretome Profiling
Objective: To collect, concentrate, and normalize conditioned media from MSCs for proteomic analysis using DIA-MS, enabling comprehensive and reproducible quantification of secreted proteins.
Materials:
- Serum-free culture medium
- Ultrafiltration centrifugal devices (e.g., 10kDa MWCO)
- BCA protein assay kit
- Trypsin/Lys-C protease mix
- StageTips or other desalting tips
- LC-MS/MS system
Method:
- Conditioned Media Collection:
- Differentiate MSCs towards desired lineages in standard differentiation media.
- Prior to secretome collection, wash cells thoroughly with PBS and incubate with serum-free medium for 24-48 hours.
- Collect conditioned medium and centrifuge at high speed to remove any dead cells and debris.
Protein Concentration and Normalization:
- Concentrate the supernatant using an ultrafiltration device. Record the initial and final volumes to calculate the Concentration Rate (CR).
- Measure protein concentration using the BCA assay as a guide.
- Normalize sample volumes based on the CR to ensure equal protein loading for digestion. For example, load a volume proportional to 1/CR for subsequent steps [118].
Protein Digestion and Peptide Clean-up:
- Reduce and alkylate proteins using DTT and iodoacetamide.
- Digest proteins into peptides overnight using Trypsin/Lys-C mix.
- Acidify peptides to stop digestion and desalt using C18 StageTips.
Mass Spectrometric Analysis (DIA):
- Reconstitute peptides in LC-MS loading buffer.
- Separate peptides on a nano-flow LC system with a C18 column gradient.
- Acquire data on a mass spectrometer capable of DIA (e.g., timsTOF, Orbitrap).
- In DIA mode, the mass spectrometer cycles through sequential, overlapping isolation windows covering a broad m/z range (e.g., 400-1000 m/z), fragmenting and analyzing all ions within each window.
Data Processing and Analysis:
- Use specialized software (e.g., Spectronaut, DIA-NN) to analyze DIA data against a spectral library generated from human protein sequences or data-dependent acquisition (DDA) runs of the same samples.
- Identify significantly differentially secreted proteins using statistical tests (e.g., t-test, ANOVA) with correction for multiple hypotheses.
Diagram 2: Experimental workflow for secretome analysis. The key normalization step based on the Concentration Rate (CR) is highlighted in red, which addresses a major methodological challenge in the field.
The Scientist's Toolkit: Essential Research Reagents and Materials
Successful execution of these protocols requires specific, high-quality reagents. The following table lists essential materials and their functions.
Table 2: Essential Research Reagents for Molecular Validation of MSC Differentiation
| Item Name | Function/Application | Key Notes |
|---|---|---|
| Osteogenic Supplements | Induce bone differentiation. | Typically contains dexamethasone, ascorbic acid, and β-glycerophosphate [44]. |
| Adipogenic Supplements | Induce fat differentiation. | Typically contains dexamethasone, indomethacin, IBMX, and insulin [44]. |
| Chondrogenic Supplements | Induce cartilage differentiation. | Often includes TGF-β (e.g., TGF-β3), dexamethasone, and ascorbic acid [44]. |
| Ultrapure RNA Kit | Isolation of high-quality RNA for qRT-PCR. | Essential for removing contaminants that inhibit downstream enzymatic reactions [117]. |
| SYBR Green qPCR Master Mix | Fluorescent detection of amplified DNA. | Contains Hot Start Taq polymerase, SYBR Green dye, dNTPs, and optimized buffer [117]. |
| Validated Primer Assays | Specific amplification of target genes. | Primers must be designed for lineage-specific markers (see Table 1) and tested for efficiency and specificity [98]. |
| Ultrafiltration Devices | Concentration of proteins from conditioned media. | Critical first step in secretome preparation; 10kDa molecular weight cut-off (MWCO) is common [118]. |
| Trypsin/Lys-C Mix | Proteolytic digestion of proteins into peptides. | Required for preparing samples for bottom-up proteomic analysis by mass spectrometry [118]. |
| C18 StageTips | Desalting and clean-up of peptide mixtures. | Removes salts and impurities that interfere with LC-MS analysis [118]. |
Application Notes and Protocols
1. Introduction Within the context of stem cell-based therapies for bone and metabolic diseases, the migratory and adhesive properties of stem cells are critical for successful homing to target tissues and subsequent engraftment. This document provides a comparative analysis of these properties in hematopoietic stem cells (HSCs) and mesenchymal stem cells (MSCs), detailing key molecular regulators, quantitative differences, and standardized protocols for investigating these processes. The insights are framed within the broader thesis research on MSC differentiation into osteogenic, chondrogenic, and adipogenic lineages.
2. Comparative Properties of Stem Cell Populations 2.1. Hematopoietic Stem Cells (HSCs) Homing and engraftment of HSCs are multistep processes dependent on specific adhesion molecules and chemokine receptors. Significant functional differences exist between short-term (ST) and long-term (LT) HSC populations.
Table 1: Comparative Adhesion and Migration Properties of Murine HSC Populations
| HSC Population | Phenotype | sLex Expression (E-selectin ligand) | CXCR4 Expression | CD26 (DPP4) Expression | Key Adhesion Deficiency | Effective Enhancement Strategy |
|---|---|---|---|---|---|---|
| Short-Term (ST) HSC | Flk2⁻CD34⁺ | >60% [120] [121] | High [120] [121] | Low [120] [121] | N/A | Fucosylation with rhFTVI [120] [121] |
| Long-Term (LT) HSC | Flk2⁻CD34⁻ | <10% [120] [121] | Low [120] [121] | High [120] [121] | Compromised E-selectin binding & CXCR4-mediated migration [120] [121] | CD26 inhibition with Diprotin A (Dip A) [120] [121] |
The low sLex expression on LT-HSCs limits their initial tethering and rolling on bone marrow endothelium, a step mediated by E-selectin [120] [122] [121]. Furthermore, lower CXCR4 expression and higher CD26 surface expression—a peptidase that deactivates the SDF-1 chemokine—compromise the subsequent SDF-1/CXCR4 signaling axis critical for transmigration and retention in the niche [120] [121].
Protocol 1: Enhancing HSC Homing and Engraftment via Molecular Engineering Objective: To improve the migration and engraftment efficiency of murine HSCs. Materials:
- Recombinant Human Fucosyltransferase VI (rhFTVI)
- Diprotin A (Dip A), a CD26 inhibitor
- Isolated murine ST-HSCs (Flk2⁻CD34⁺) and LT-HSCs (Flk2⁻CD34⁻) Method:
- HSC Isolation: Isolate ST-HSCs and LT-HSCs from mouse bone marrow mononuclear cells via fluorescence-activated cell sorting (FACS) using a lineage cocktail (CD5, CD11b, CD45R, anti–7-4, anti–Gr-1, anti–terr-119) and antibodies against Sca-1, c-Kit, CD34, and Flk2 [120].
- Pre-treatment:
- Transplantation: Transplant treated cells into recipient mice via intravenous injection.
- Engraftment Analysis: Monitor long-term engraftment levels in bone marrow. Fucosylated ST-HSCs show enhanced capacity to engraft secondary recipients, while Dip A-treated LT-HSCs show significantly improved primary engraftment [120] [121].
2.2. Mesenchymal Stem Cells (MSCs) The adhesion and migration of MSCs are regulated by integrins, focal adhesion dynamics, and key extracellular matrix (ECM) components, which also influence lineage commitment.
Table 2: Key Molecular Regulators in MSC Adhesion, Migration, and Differentiation
| Molecule | Function/Role | Effect on Osteogenesis | Effect on Adipogenesis | Associated Pathway/Process |
|---|---|---|---|---|
| Lamc1 (Laminin subunit gamma-1) | ECM component | Promotes [98] | Inhibits [98] | ECM-receptor interaction, Focal adhesion [98] |
| Integrin β1 | Adhesion receptor | Promotes [123] [98] | Inhibits [98] | Integrin-mediated signaling, FAK activation [123] [72] |
| Cytohesin 1 (CYTH1) | Adhesion regulator | Implicated in homing (via integrin β1 activation) [123] | Not specified | Integrin activation, Homing [123] |
| ERK (MAPK) | Kinase | Promotes [29] | Inhibits (inhibition redirects to adipogenesis) [29] | Mitogen-activated Protein Kinase (MAPK) signaling [29] |
| p38 (MAPK) | Kinase | Promotes (late differentiation) [29] | Not specified | MAPK signaling [29] |
| Substrate Rigidity | Biophysical cue | Favored on stiffer substrates [124] | Not specified | Mechanotransduction [124] |
Lamc1, a laminin subunit, is a critical ECM component that promotes osteogenic differentiation while inhibiting adipogenic differentiation in BMSCs. Knockdown of Lamc1 inhibits both lineages [98]. The ERK pathway is a key switch; its sustained activation directs MSCs toward osteogenesis, while its inhibition blocks osteogenesis and promotes adipogenesis [29].
Protocol 2: Investigating MSC Migration on Dynamic Soft Substrates Objective: To assess MSC migration under conditions of rapidly switching substrate rigidity. Materials:
- Photo-responsive yellow protein (PYP) hydrogels (Young's modulus switchable between ~1.6 kPa and ~2.2 kPa)
- Polyacrylamide (PA) hydrogels (static control, ~2.2 kPa)
- Time-lapse microscopy system Method:
- Cell Seeding: Culture human MSCs (hMSCs) on PYP hydrogels and allow them to equilibrate for 24 hours.
- Rigidity Cycling: Subject the experimental group to cyclic illumination (e.g., 1-minute light/1-minute dark intervals) for 12 hours to induce fast, reversible rigidity changes. Maintain control groups in static dark (modulus ~2.2 kPa) or static light (~1.6 kPa) conditions.
- Image Acquisition and Analysis: Record cell movements using time-lapse microscopy. Quantify migration speed and observe morphological changes.
- Expected Outcome: hMSCs on dynamic soft substrates can exhibit a 36-fold increase in migration speed compared to static soft substrates, accompanied by periodic elongation and "snap-back" morphological changes, not seen in traditional mesenchymal migration [124].
3. The Scientist's Toolkit: Essential Reagents and Materials
Table 3: Research Reagent Solutions for Homing and Adhesion Studies
| Reagent/Material | Function/Application | Key Example/Benefit |
|---|---|---|
| Recombinant Human Fucosyltransferase VI (rhFTVI) | Enzymatically fucosylates cell surface proteins to enhance E-selectin ligand (sLex) expression. | Improves homing and engraftment of ST-HSCs [120] [121]. |
| Diprotin A (Dip A) | Inhibits cell surface CD26 (DPP4) peptidase activity, protecting SDF-1 from degradation. | Enhances CXCR4-mediated migration and engraftment of LT-HSCs [120] [121]. |
| Photo-responsive PYP Hydrogels | Substrates with dynamically and reversibly switchable rigidity in response to light. | Enables study of cell migration under dynamic mechanical cues [124]. |
| Polarized PVDF or PLLA Scaffolds | Piezoelectric biomaterials that generate electrical signals in response to mechanical stress. | Enhances stem cell adhesion strength and focal adhesion maturation [72]. |
| PD98059 | Specific inhibitor of MEK-1, the upstream kinase of ERK. | Used to inhibit ERK signaling and study its role in osteogenic vs. adipogenic lineage commitment [29]. |
4. Signaling Pathways Regulating MSC Fate and Homing The following diagrams illustrate the core signaling pathways and experimental workflows discussed.
Diagram 1: Key pathways in MSC fate decision. This diagram illustrates how extracellular matrix (ECM) components and adhesion signals, including Lamc1 and Integrin β1, converge on focal adhesion kinase (FAK) and ERK/MAPK signaling to promote osteogenic differentiation while inhibiting adipogenic differentiation [29] [98].
Diagram 2: HSC homing enhancement strategy. This workflow outlines the distinct pre-treatment strategies for short-term (ST) and long-term (LT) HSCs to overcome their specific molecular deficiencies in the homing process, leading to improved engraftment [120] [121].
5. Conclusion The migratory and adhesive properties of HSCs and MSCs are fundamentally regulated by distinct but overlapping sets of adhesion molecules, signaling pathways, and biophysical cues. Understanding these differences is paramount for developing targeted strategies to enhance the efficacy of stem cell therapies in regenerative medicine, particularly in the context of directing MSC lineage commitment for skeletal and adipose tissue repair. The protocols and reagents detailed herein provide a foundation for advanced pre-clinical research in this field.
Within the broader context of stem cell differentiation research, understanding the repair of musculoskeletal tissues is paramount. Mesenchymal stem cells (MSCs), with their capacity for osteogenic, adipogenic, and chondrogenic differentiation, play a central role in the healing of bone and cartilage [5]. In vivo models are indispensable for deciphering the complex signaling pathways that govern MSC fate and for translating promising in vitro findings into clinically effective therapies. These models provide the necessary physiological environment to study the interplay between scaffolds, cells, and signaling molecules, thereby bridging the gap between basic stem cell biology and regenerative medicine applications [125]. This document provides a detailed overview of the predominant animal models used to assess bone and cartilage repair, with a specific focus on their application in stem cell-driven research.
Animal Models for Bone Defect Repair
Preclinical models for bone repair must replicate the clinical challenge of critical-sized defects (CSDs)—those that will not heal spontaneously without intervention. The choice of model is critical and depends on the specific research question, whether it is screening new biomaterials or studying the fundamental biology of healing.
Table 1: Key Animal Models for Bone Defect Research
| Model Type | Species | Defect Location | Healing Mechanism | Key Advantages | Primary Translational Limitations |
|---|---|---|---|---|---|
| Calvarial Defect | Rat, Mouse | Skull | Intramembranous Ossification [126] | High reproducibility, cost-effective, minimal fixation needed, suitable for transgenic studies [126] | Heals via intramembranous pathway only; lacks biomechanical loading [126] |
| Drill-Hole Defect | Various | Long Bone Metaphysis/Epiphysis | Trabecular bone healing via direct membranous formation; cortical healing involves both endochondral and intramembranous ossification [127] | Excellent for studying trabecular bone regeneration; highly standardized; no external fixation required [127] | Less directly translational to clinical shaft fractures; defect size is small [127] |
| Critical-Size Long Bone Defect | Rabbit, Minipig, Sheep | Femur, Tibia, Radius | Endochondral Ossification [126] | Clinically relevant healing pathway (endochondral); suitable for testing under load-bearing conditions [126] | Technically challenging; requires robust fixation; high costs and ethical considerations [126] |
Detailed Protocol: Rodent Calvarial Defect Model
The rodent calvarial defect model (RCD) is a widely used platform for the initial evaluation of osteoconductive biomaterials and stem cell-based constructs [126].
Experimental Workflow: The following diagram outlines the key stages in establishing and analyzing a calvarial defect model.
Key Reagents and Materials:
- Animals: Adult Sprague-Dawley or Wistar rats (typically 8-12 weeks old) are commonly used. The age must be standardized as healing capacity is age-dependent [126].
- Critical-Size Defect (CSD) Tool: A trephine drill bit (5-8 mm diameter) is used to create a full-thickness defect in the parietal bone. An 8 mm defect is widely accepted as a CSD in adult rats [126].
- Test Material: This could be a biomaterial scaffold (e.g., the GelMA/DBM/DFO hydrogel [128]), often seeded with MSCs or osteoprogenitor cells.
- Fixative: 4% Paraformaldehyde (PFA) for sample preservation post-harvest.
- Analysis Reagents: Specific antibodies for immunohistochemistry (IHC), such as anti-Bone Morphogenetic Protein-2 (BMP-2) and anti-Collagen Type I (COL-I), are used to confirm osteogenic differentiation and new bone matrix synthesis [128].
Animal Models for Cartilage Lesion Repair
Cartilage repair is challenging due to its avascular nature. Models range from small rodents, ideal for genetic studies, to large animals, which better mimic human joint mechanics.
Table 2: Key Animal Models for Cartilage Lesion Research
| Model Type | Species | Lesion Characteristics | Key Advantages | Primary Translational Limitations |
|---|---|---|---|---|
| Full-Thickness Articular Defect (Small) | Mouse, Rat | Defined width (0.1-0.3 mm) and depth (to subchondral bone) [129] | Cost-effective; enables use of genetically modified strains; high throughput for mechanistic studies [129] | Small size makes intervention challenging; high intrinsic repair capacity in some strains [129] |
| Osteochondral Defect (Large) | Minipig, Goat, Sheep | Larger defects (e.g., 8-10 mm) in femoral condyle or trochlea [130] | Joint size and cartilage thickness closely resemble humans; suitable for testing cell-based implants like allogenic cartilage beads [130] | High costs, specialized housing, and ethical constraints; limited availability of species-specific reagents |
| Allogenic Implant Model | Minipig | Implantation of hyaline-like bioengineered cartilage minigrafts (e.g., Cartibeads) into chondral lesions [130] | Demonstrates safety and efficacy of off-the-shelf products; avoids donor-site morbidity; one-step surgery [130] | Requires specialized production facilities; potential for immune response (though minimal with chondrocytes) [130] |
Detailed Protocol: Minipig Osteochondral Defect Model
This protocol describes the implantation of allogenic bioengineered cartilage beads, a promising advanced therapy, in a large animal model [130].
Experimental Workflow: The diagram below illustrates the key steps from the preparation of allogenic implants to the final in vivo assessment.
Key Reagents and Materials:
- Animals: Göttingen minipigs, aged 20-28 months, are used for their cartilage maturity and knee anatomy similarity to humans [130].
- Bioengineered Cartilage Beads: Allogenic minigrafts (e.g., Cartibeads) produced via a patented method involving dedifferentiation and redifferentiation of donor chondrocytes to form hyaline-like cartilage [130].
- Surgical Tools: Cylindrical punch biopsy tool and curette for creating defined chondral lesions without damaging the subchondral bone.
- Surgical Adhesive: Fibrin-based surgical glue (e.g., Tisseel) to secure the implanted beads within the lesion [130].
- Analysis Tools: Magnetic resonance imaging (MRI) for non-invasive assessment of tissue coverage and integration, and histological grading systems (e.g., Bern score) for evaluating the quality of the repaired tissue [130].
The Scientist's Toolkit: Key Research Reagents
The following table lists essential reagents and materials frequently used in bone and cartilage regeneration studies, as highlighted in the cited research.
Table 3: Essential Research Reagents for Bone and Cartilage Repair Studies
| Reagent/Material | Function and Application | Example from Research |
|---|---|---|
| Demineralized Bone Matrix (DBM) | An allogeneic bone graft providing a collagenous scaffold and native growth factors (e.g., BMPs) to support osteogenesis [128]. | Used in a composite GelMA/DBM/DFO hydrogel to promote cranial defect repair in rats [128]. |
| Gelatin Methacrylate (GelMA) | A photopolymerizable hydrogel that provides a hydrating, ECM-mimetic 3D environment for cell encapsulation and tissue growth [128]. | Serves as the base material in composite hydrogels for bone repair, allowing modular incorporation of DBM and bioactive factors [128]. |
| Deferoxamine (DFO) | A hypoxia-mimetic agent that stabilizes HIF-1α, upregulating angiogenic factors to promote vascularization, which is crucial for bone regeneration [128]. | Loaded into GelMA/DBM hydrogels to enhance angiogenesis and improve bone repair outcomes in a rat model [128]. |
| Reactive Oxygen Species (ROS) Modulators | Chemicals to manipulate intracellular ROS levels, which are key signaling molecules influencing MSC lineage commitment toward adipogenesis or osteogenesis [131]. | Used in vitro to demonstrate that elevated ROS levels promote adipogenic differentiation of MSCs via the CHOP-Akr1A1 pathway [131]. |
| Functionalized Acrylate Polymers | Synthetic substrates (e.g., grafted with gelatin or heparin) used to study the impact of extracellular matrix components on MSC adhesion, proliferation, and differentiation [6]. | Used in 2D and 3D cultures to demonstrate that substrate biochemistry can direct spontaneous osteogenic commitment of MSCs [6]. |
Signaling Pathways in Mesenchymal Stem Cell Differentiation
Understanding the molecular pathways that dictate MSC fate is fundamental to developing targeted regenerative strategies. The following diagram summarizes key pathways involved in osteogenic and chondrogenic differentiation, as identified in the in vivo studies.
Pathway Insights:
- ROS/CHOP/Akr1A1 Pathway: Elevated reactive oxygen species (ROS) levels increase the expression of transcription factor CHOP, which in turn upregulates Aldo-keto reductase family 1 member A1 (Akr1A1). High Akr1A1 expression inhibits the SIRT1/PGC-1α/TAZ axis, shifting MSC differentiation toward adipogenesis at the expense of osteogenesis [131]. This pathway is significant in age-related and metabolic bone loss.
- PTH/PTHrP and Hedgehog Signaling: In a mouse cartilage repair model, both the PTH/PTHrP and Hedgehog (Hh) signaling pathways were activated during the repair process. These pathways converge on transcription factors like CREB (cAMP response element-binding protein), promoting the expression of genes essential for osteogenic and chondrogenic differentiation [129].
In regenerative medicine and tissue engineering, the successful translation of laboratory innovations into clinical therapies hinges on their performance against the established clinical gold standard: the autologous graft. Autologous grafts, which involve transplanting a patient's own tissues from one site to another, represent the benchmark due to their inherent biocompatibility and viability. However, their use is constrained by significant limitations, including donor-site morbidity, limited tissue availability, and the need for additional surgical procedures. This application note details the critical protocols and quantitative benchmarks for comparing novel tissue-engineered constructs to autologous grafts, with a specific focus on applications informed by stem cell differentiation research into osteogenic, chondrogenic, and adipogenic lineages.
Quantitative Benchmarking of Performance Outcomes
Rigorous preclinical and clinical studies provide the primary data for comparing new constructs to autologous grafts. The following tables summarize key quantitative outcomes from recent investigations in vascular and neural regeneration, two fields where benchmarking is advanced.
Table 1: Benchmarking a Bioengineered Vascular Conduit against Autologous Vein Graft [132]
| Outcome Measure | Symvess ATEV Performance | Autologous Vein Graft Performance | Statistical Significance (p-value) |
|---|---|---|---|
| Patency | No significant difference | No significant difference | Not Significant (NS) |
| Amputation Rate | No significant difference | No significant difference | NS |
| Infection Rate | No significant difference | No significant difference | NS |
| Reintervention Rate | No significant difference | No significant difference | NS |
| Conduit Complication | No significant difference | No significant difference | NS |
| Death | No significant difference | No significant difference | NS |
Key Insight: The acellular tissue-engineered vessel (ATEV) demonstrated performance statistically indistinguishable from autologous vein across all major clinical outcome measures in a propensity-matched study, suggesting its potential as a viable alternative when autologous vein is unsuitable [132].
Table 2: Preclinical Performance of Natural Material-Based Nerve Guidance Conduits (NGCs) [133]
| Conduit Material | Experimental Model | Key Differentiation & Functional Outcomes | Benchmark against Autograft |
|---|---|---|---|
| Collagen-based (NeuraGen 3D + Schwann Cells) | Long-gap rat model | Supported axonal regeneration | Regeneration comparable to autograft |
| Collagen-based + AGRG Hydrogel | Chronic 25 mm gap in rabbits | Positive histological and functional recovery | Outcomes similar to autograft |
| Chitosan-Collagen Conduit | Not specified | Superior functional recovery | Superior to silicone conduits; performance relative to autograft not specified |
| Avance Decellularized Allograft + MSCs | In vitro seeding study | High MSC viability and uniform distribution | N/A (Structure/function study) |
Table 3: Osteogenic and Chondrogenic Potential of MSCs from Different Tissue Sources [134] [135]
| Cell Source | Osteogenic Differentiation Markers | Chondrogenic Differentiation Markers | Inferred Potential vs. Bone Marrow |
|---|---|---|---|
| Bone Marrow-derived MSCs (BMMSCs) | High Alkaline Phosphatase (AP) activity; significant matrix mineralization [134] | High histological score (6.5 ± 1.3); robust matrix production [134] | Gold Standard |
| Adipose-derived MSCs (ATMSCs) | Significantly less AP activity and mineralization vs. BMMSC (p=0.002) [134] | Inferior histological score (4.3 ± 1.6) vs. BMMSC (p=0.023) [134] | Inferior |
| Buccal Fat Pad-derived MSCs (BFP-MSCs) | 7x105-fold increase in BGLA mRNA; 733-fold increase in BMP2 mRNA [135] | 282-fold higher expression of Collagen I mRNA [135] | High (in study context) |
| Gingiva-derived Cells (GDCs) | Fewer mineralized nodules; no significant mRNA increase in markers [135] | Slight morphological transformation; no significant mRNA increase in Collagen I [135] | Lower |
Detailed Experimental Protocols for Benchmarking
Protocol: In Vivo Benchmarking of a Vascular Conduit
This protocol is adapted from a study comparing the Symvess ATEV to autologous vein grafts for vascular repair [132].
1. Objective: To evaluate the safety and efficacy of a tissue-engineered vascular conduit against the autologous vein graft standard in a clinically relevant model.
2. Materials:
- Test Article: Acellular tissue-engineered vessel (e.g., Symvess).
- Control Article: Autologous vein graft harvested from the subject.
- Animal Model: Appropriate large animal model (e.g., porcine, canine).
- Surgical Equipment: Standard microvascular surgical set.
- Analysis Tools: Histology, immunohistochemistry, ultrasound for patency monitoring.
3. Methodology: 1. Study Design: Conduct a prospective, randomized study or a propensity-matched analysis of clinical/historical data. 2. Implantation: Implant the test and control conduits in anatomically comparable positions. 3. Outcome Monitoring: * Primary Patency: Assess via Doppler ultrasound or angiography at predefined endpoints. * Morbidity: Monitor for amputation, infection, and other conduit-related complications. * Histological Analysis: Explant conduits at study termination. Process for H&E staining, and immunohistochemical staining for endothelial cell markers (e.g., CD31) and smooth muscle cell markers (e.g., α-SMA) to assess cellular repopulation and tissue remodeling [132]. 4. Statistical Analysis: Compare outcomes between groups using appropriate statistical tests (e.g., log-rank test for patency, chi-square for complication rates). The study should be powered to detect non-inferiority.
Protocol: In Vitro Benchmarking of MSC Differentiation Potential
This protocol outlines the standard methods for comparing the osteogenic and chondrogenic capacity of MSCs from different sources, a critical step in selecting cells for engineered tissues [134] [135].
1. Objective: To compare the osteogenic and chondrogenic differentiation potential of two populations of Mesenchymal Stem/Stromal Cells (e.g., bone marrow-derived vs. adipose-derived).
2. Materials:
- Cell Sources: BMMSCs and ATMSCs.
- Culture Media: Basal medium (e.g., DMEM), growth medium (with FBS), osteogenic induction medium (containing dexamethasone, β-glycerophosphate, and ascorbate), chondrogenic induction medium (containing TGF-β and IGF-I).
- Staining Reagents: Alizarin Red S or Von Kossa stain for mineralization; Alkaline Phosphatase (AP) stain; Safranin-O for proteoglycans.
- Molecular Biology Reagents: RNA isolation kit, cDNA synthesis kit, qPCR reagents, primers for osteogenic (e.g., BGLA, BMP2) and chondrogenic (e.g., COLL) markers.
3. Methodology: * A. Osteogenic Differentiation (Monolayer Culture): 1. Seed cells at a defined density in multi-well plates. 2. At 60-80% confluence, replace growth medium with osteogenic induction medium. Maintain control cells in growth medium. 3. Culture for 2-3 weeks, changing the medium twice weekly. 4. Analysis: * Cellular Staining: Fix cells and perform AP staining at an intermediate time point (e.g., 7-14 days) and Alizarin Red S or Von Kossa staining at terminal points to visualize calcium deposits. * Molecular Analysis: Extract RNA at multiple time points. Use qRT-PCR to quantify expression of osteogenic genes like BGLA and BMP2 [135].
Signaling Pathways and Experimental Workflows
The differentiation of MSCs is governed by complex and often reciprocal signaling pathways. Understanding these is key to engineering tissues that can rival autografts.
Diagram 1: Signaling pathways and reciprocal inhibition in MSC differentiation. Pathways are activated by specific induction factors. A critical concept is the reciprocal inhibition between osteogenic and adipogenic lineages; stimulating one often suppresses the other, a balance important for bone homeostasis [28].
Diagram 2: A generalized workflow for the in vivo benchmarking of a tissue-engineered construct (e.g., vascular conduit or nerve guide) against an autologous graft.
The Scientist's Toolkit: Research Reagent Solutions
Table 4: Essential Reagents for MSC Differentiation and Tissue Engineering Studies
| Reagent / Material | Function / Application | Example in Context |
|---|---|---|
| Induction Media | Directs stem cell fate by providing specific biochemical cues. | Osteogenic medium (Dexamethasone, β-glycerophosphate, Ascorbate) [134]; Chondrogenic medium (TGF-β, IGF-I) [134]. |
| Flow Cytometry Antibodies | Characterizes and purifies MSC populations based on surface markers. | Positive markers: CD73, CD90, CD105, CD44 [28] [135]. Negative markers: CD34, CD45 [28] [135]. |
| Histological Stains | Visualizes differentiation outcomes and extracellular matrix production. | Alizarin Red / Von Kossa (calcium deposits); Safranin-O (proteoglycans); Immunostaining for Collagen Type II [134] [135]. |
| qRT-PCR Primers | Quantifies mRNA expression of lineage-specific genes. | Primers for BGLA, BMP2 (osteogenesis), COLL (chondrogenesis) [135]. |
| 3D Scaffolds & Conduits | Provides structural support and biomechanical cues for tissue formation. | Collagen-based conduits (NeuraGen) [133]; Chitosan scaffolds [133]; Fibrin gels for pellet culture [134]. |
| Decellularized Allografts | Provides a natural, bioinstructive ECM scaffold for implantation. | Avance nerve graft; can be seeded with MSCs ex vivo [133]. |
Conclusion
The directed differentiation of MSCs into osteogenic, chondrogenic, and adipogenic lineages represents a cornerstone of modern regenerative medicine. This synthesis of foundational science, advanced methodologies, and rigorous validation underscores a powerful convergence of biology and engineering. Future progress hinges on overcoming key challenges in standardization, scalability, and faithful recapitulation of in vivo conditions. Emerging technologies, particularly in AI-driven prediction, sophisticated biomaterials, and gene editing, are poised to unlock unprecedented control over stem cell fate. The continued translation of these insights promises to revolutionize the treatment of bone fractures, osteoarthritis, osteoporosis, and other debilitating conditions, ultimately enabling the development of robust, off-the-shelf regenerative therapies.
References
- 1. Mesenchymal stem cells in treating human diseases [https://www.nature.com/articles/s41392-025-02313-9]
- 2. Mesenchymal stem cells: opening a new chapter in the ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12465317/]
- 3. Validated methods for isolation and qualification of ... [https://translational-medicine.biomedcentral.com/articles/10.1186/s12967-025-06972-8]
- 4. Mesenchymal Stem Cell-Based Therapies: Challenges ... [https://link.springer.com/article/10.1007/s12013-025-01895-z]
- 5. Adipogenesis, Osteogenesis, and Chondrogenesis of ... [https://www.frontiersin.org/journals/cell-and-developmental-biology/articles/10.3389/fcell.2020.00561/full]
- 6. Maintenance of pluripotency and osteogenic differentiation ... [https://www.nature.com/articles/s41598-025-05640-7]
- 7. XYLT1 Deficiency of Human Mesenchymal Stem Cells [https://pubmed.ncbi.nlm.nih.gov/40806493/]
- 8. Mesenchymal stromal cell therapy: Progress to date and ... [https://www.sciencedirect.com/science/article/pii/S1525001625000930]
- 9. Transcriptional Regulatory Cascades in Runx2-Dependent ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3627420/]
- 10. Transcription Factor Osterix - an overview [https://www.sciencedirect.com/topics/medicine-and-dentistry/transcription-factor-osterix]
- 11. Wnt/β-catenin signaling components and mechanisms in ... [https://www.nature.com/articles/s41413-024-00342-8]
- 12. Crosstalk between Wnt and bone morphogenetic protein ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10915952/]
- 13. The Smad Dependent TGF-β and BMP Signaling Pathway ... [https://www.frontiersin.org/journals/molecular-biosciences/articles/10.3389/fmolb.2021.593310/full]
- 14. RUNX2-modifying enzymes: therapeutic targets for bone ... [https://www.nature.com/articles/s12276-020-0471-4]
- 15. regulated Interactions between Osterix and Runx2 Are ... [https://www.sciencedirect.com/science/article/pii/S0021925820371684]
- 16. Osterix represses adipogenesis by negatively regulating ... [https://www.nature.com/articles/srep35655]
- 17. Wnt/β-catenin signaling pathway: proteins' roles in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11520425/]
- 18. Stimulating factors for regulation of osteogenic and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10277964/]
- 19. Sox9 Determines Translational Capacity During Early ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8256280/]
- 20. Regulation and function of SOX9 during cartilage ... [https://www.sciencedirect.com/science/article/abs/pii/S1044579X20300961]
- 21. Dominant role of in situ native cartilage niche for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8843973/]
- 22. Article Cross-Regulation of C/EBPα and PPARγ Controls ... [https://www.sciencedirect.com/science/article/pii/S1097276500803068]
- 23. PPARγ and C/EBP factors orchestrate adipocyte biology via ... [https://genesdev.cshlp.org/content/22/21/2941.full.html]
- 24. C/EBPα induces adipogenesis through PPARγ: a unified ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC155311/]
- 25. C/EBPα induces adipogenesis through PPARγ [https://genesdev.cshlp.org/content/16/1/22]
- 26. Prmt6 represses the pro-adipogenic Ppar-gamma–C/ebp ... [https://www.nature.com/articles/s41598-024-57310-9]
- 27. Enhancing adipogenesis in Wharton's jelly multipotent ... [https://www.nature.com/articles/s41598-025-16867-9]
- 28. Adipogenesis, Osteogenesis, and Chondrogenesis of Human ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7362937/]
- 29. Adult Human Mesenchymal Stem Cell Differentiation to the ... [https://www.sciencedirect.com/science/article/pii/S0021925818300838]
- 30. Fate decision of mesenchymal stem cells: adipocytes or ... [https://www.nature.com/articles/cdd2015168]
- 31. A novel PPARγ2 modulator sLZIP controls the balance ... [https://www.nature.com/articles/cdd201480]
- 32. Mapk7 enhances osteogenesis and suppresses ... [https://www.nature.com/articles/s42003-025-07765-x]
- 33. Determinants of stem cell lineage differentiation toward ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6456412/]
- 34. Signaling Pathways Driving MSC Osteogenesis [https://www.mdpi.com/1422-0067/26/3/1311]
- 35. Omics characteristics and epigenetic modifications of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12590281/]
- 36. Comparative characterization and osteogenic / adipogenic ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7417469/]
- 37. Osteogenic shift in the adipose-derived stem cells of ... [https://www.frontiersin.org/journals/cell-and-developmental-biology/articles/10.3389/fcell.2025.1603405/full]
- 38. Surface topography enhances differentiation of ... [https://www.sciencedirect.com/science/article/abs/pii/S0142961215004809]
- 39. Dynamics of lineage commitment revealed by single-cell ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5653659/]
- 40. Dynamic Transcriptome Profiling Reveals LncRNA ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9130768/]
- 41. Transcriptomic analysis of long non coding RNAs and their ... [https://www.nature.com/articles/s41598-025-14660-2]
- 42. Single-Cell Transcriptomic Analysis Defines Heterogeneity ... [https://www.sciencedirect.com/science/article/pii/S2211124716317673]
- 43. High-resolution spatial transcriptomics and cell lineage ... [https://www.nature.com/articles/s41467-025-59206-2]
- 44. Protocols for in vitro Differentiation of Human Mesenchymal ... [https://pubmed.ncbi.nlm.nih.gov/27236670/]
- 45. Identification of potential specific biomarkers and key ... [https://josr-online.biomedcentral.com/articles/10.1186/s13018-020-01965-3]
- 46. Scientists discover new way to shape what a stem cell ... [https://www.colorado.edu/today/2025/11/03/scientists-discover-new-way-shape-what-stem-cell-becomes]
- 47. Small molecules for mesenchymal stem cell fate ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6904864/]
- 48. Recent advances on small molecules in osteogenic ... [https://stemcellres.biomedcentral.com/articles/10.1186/s13287-022-03204-4]
- 49. Potential mechanisms and applications of statins on ... [https://www.sciencedirect.com/science/article/abs/pii/S0168365915300341]
- 50. Fluvastatin promotes chondrogenic differentiation of adipose ... [https://bmcpharmacoltoxicol.biomedcentral.com/articles/10.1186/s40360-022-00600-7]
- 51. Adenosine Signaling Mediates Osteogenic Differentiation ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4639610/]
- 52. Metformin Promotes Osteogenic Differentiation of Adipose ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6306720/]
- 53. Osteogenic Potential of Simvastatin and Fluvastatin in an ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12299731/]
- 54. Metformin potentiates immunosuppressant activity and ... [https://www.sciencedirect.com/science/article/pii/S1567576923014042]
- 55. Maintenance of pluripotency and osteogenic differentiation ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12217605/]
- 56. Gelatin-based biomaterials and gelatin as an additive for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11094280/]
- 57. Fabrication of gelatin-heparin based cartilage models [https://www.frontiersin.org/journals/biomaterials-science/articles/10.3389/fbiom.2024.1331032/full]
- 58. Chemical group-dependent plasma polymerisation ... [https://www.sciencedirect.com/science/article/pii/S1742706116306845]
- 59. A 3D Self-Assembly Platform Integrating Decellularized ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12048290/]
- 60. Recapitulating the bone extracellular matrix through 3D ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12177472/]
- 61. Development of a 3D cell-printed RVO model by advancing ... [https://link.springer.com/article/10.1007/s42114-025-01455-2]
- 62. Optimizing extracellular matrix for endothelial ... [https://www.nature.com/articles/s41598-025-09256-9]
- 63. Application of decellularization methods for scaffold ... [https://www.frontiersin.org/journals/bioengineering-and-biotechnology/articles/10.3389/fbioe.2025.1621641/full]
- 64. Design and Applications of Extracellular Matrix Scaffolds in ... [https://www.mdpi.com/2073-4409/14/14/1076]
- 65. 3D bioprinting approaches for musculoskeletal interfaces ... [https://www.sciencedirect.com/science/article/pii/S0378517325007768]
- 66. The control of stem cell morphology and differentiation ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6561507/]
- 67. Application of Machine Learning in Predicting Osteogenic ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12561851/]
- 68. Quantification of the morphological characteristics of hESC ... [https://www.nature.com/articles/s41598-019-53719-9]
- 69. Early and non-destructive prediction of the differentiation ... [https://www.nature.com/articles/s41598-025-11108-5]
- 70. A new protocol for validation of Chondro, Adipo and Osteo ... [https://www.sciencedirect.com/science/article/abs/pii/S0040816622001951]
- 71. Adipogenic, osteogenic, and chondrogenic differentiation [https://bio-protocol.org/exchange/minidetail?id=614722&type=30]
- 72. Bioelectric cues from piezoelectric materials in stem-cell ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12631758/]
- 73. Open Challenges and Opportunities in Piezoelectricity for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12520560/]
- 74. Biodegradable Piezoelectric Zinc Oxide Composite ... [https://pubmed.ncbi.nlm.nih.gov/41024652/]
- 75. Ceramic-based piezoelectric material reinforced 3D printed ... [https://www.sciencedirect.com/science/article/pii/S0264127525009621]
- 76. A comparative analysis of stem cells derived from young ... [https://www.frontiersin.org/journals/cell-and-developmental-biology/articles/10.3389/fcell.2025.1539308/full]
- 77. Piezoelectric effect stimulates the rearrangement of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9079777/]
- 78. Electrically-stimulated cellular and tissue events are ... [https://www.sciencedirect.com/science/article/abs/pii/S0142961224003880]
- 79. Tensile stress promotes the chondrogenic ability of condylar ... [https://stemcellres.biomedcentral.com/articles/10.1186/s13287-025-04439-7]
- 80. Chondrogenic and Osteogenic In Vitro Differentiation ... [https://www.mdpi.com/2077-0383/14/4/1184]
- 81. Donor age and breed determine mesenchymal stromal cell ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11871689/]
- 82. Deciphering influence of donor age on adipose-derived ... [https://www.nature.com/articles/s41598-024-73875-x]
- 83. Age-Related Effects on the Potency of Human Adipose ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4241956/]
- 84. Donor age effects on in vitro chondrogenic and osteogenic ... [https://bmcvetres.biomedcentral.com/articles/10.1186/s12917-022-03475-2]
- 85. Comparison of biological properties of human adipose ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12574098/]
- 86. Donor Variability and 3D Culture Models Influence Human ... [https://pubmed.ncbi.nlm.nih.gov/40407303/]
- 87. Considerations for high-yield, high-throughput cell ... [https://www.nature.com/articles/s41598-018-36698-1]
- 88. Strategies for Isolating and Enriching Cancer Stem Cells [https://pmc.ncbi.nlm.nih.gov/articles/PMC3730373/]
- 89. Exploration of biomaterial and stem cell-based strategies for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12574274/]
- 90. A literature review: machine learning-based stem cell ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11193562/]
- 91. Top 10 Tips for Data Standardization Success in 2025 [https://www.intellectyx.com/top-10-best-practices-for-effective-data-standardization-in-2025/]
- 92. Data Standardization: The Ultimate Guide to Why It Matters ... [https://improvado.io/blog/data-standardization-guide]
- 93. Interpretable deep learning of single-cell and epigenetic ... [https://www.nature.com/articles/s41598-025-89646-1]
- 94. Morphology-based deep learning approach for predicting ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10720363/]
- 95. Data standardization: Why and how to standardize data [https://www.rudderstack.com/blog/data-standardization/]
- 96. Advances and continuing challenges in differentiation of ... [https://pubmed.ncbi.nlm.nih.gov/41214197/]
- 97. How the mechanical microenvironment of stem cell growth ... [https://stemcellres.biomedcentral.com/articles/10.1186/s13287-022-03070-0]
- 98. Lamc1 promotes osteogenic differentiation and inhibits ... [https://www.nature.com/articles/s41598-024-69629-4]
- 99. recent developments and future prospects in stem-cell therapy [https://pmc.ncbi.nlm.nih.gov/articles/PMC11634165/]
- 100. Stem cell therapy: a revolutionary cure or a pandora's box [https://stemcellres.biomedcentral.com/articles/10.1186/s13287-025-04334-1]
- 101. Scaling up pluripotent stem cell-based therapies [https://www.sciencedirect.com/science/article/pii/S1465324925006784]
- 102. Addressing Performance, Scalability, and Regulatory ... [https://www.biopharminternational.com/view/addressing-performance-scalability-and-regulatory-challenges-to-accelerate-cell-therapy-manufacturing]
- 103. Scalable manufacturing of clinical‐grade differentiated ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9357358/]
- 104. Current Landscape of FDA Stem Cell Approvals and Trials ... [https://www.reprocell.com/blog/current-landscape-of-fda-stem-cell-approvals-and-trials-2023-2025]
- 105. Recent Advances in Enhancement Strategies for Osteogenic ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8904963/]
- 106. Osteogenic potential: Comparison between bone marrow ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4131270/]
- 107. Adipose-derived and bone marrow mesenchymal stem cells [https://stemcellres.biomedcentral.com/articles/10.1186/s13287-018-0914-1]
- 108. Promoting Osteogenic Differentiation of Human Adipose ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6636464/]
- 109. In-vitro chondrogenic potential of synovial stem cells and ... [https://pubmed.ncbi.nlm.nih.gov/28124102/]
- 110. Rejuvenation of chondrogenic potential in a young stem ... [https://www.sciencedirect.com/science/article/abs/pii/S014296121301199X]
- 111. Exploring In Vitro Mesenchymal Stem Cell ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12408755/]
- 112. Integration of stem cells, hydrogels, and microspheres [https://www.sciencedirect.com/science/article/pii/S1385894725014147]
- 113. FTO-mediated MMP1 m6A modification promotes ... [https://www.nature.com/articles/s41598-025-13044-w]
- 114. Identification of a Novel Gene ARNT2 for Osteogenic ... [https://link.springer.com/article/10.1007/s00223-025-01407-4]
- 115. The potential application of concentrated growth factor in ... [https://stemcellres.biomedcentral.com/articles/10.1186/s13287-019-1247-4]
- 116. Identification and Validation of a New Functional Gene ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12626687/]
- 117. Selection and evaluation of reference genes for qRT-PCR ... [https://www.frontiersin.org/journals/microbiology/articles/10.3389/fmicb.2025.1500043/full]
- 118. A robust protocol for proteomic profiling of secreted ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12461699/]
- 119. Secretome Analysis: Reading Cellular Sign Language to ... [https://www.sciencedirect.com/science/article/pii/S1535947623002037]
- 120. Refining the migration and engraftment of short-term and long-term HSCs by enhancing homing-specific adhesion mechanisms - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC9636332/]
- 121. Refining the migration and engraftment of short-term and long-term HSCs by enhancing homing-specific adhesion mechanisms - PubMed [https://pubmed.ncbi.nlm.nih.gov/35764498/]
- 122. Homing and Engraftment of Hematopoietic Stem Cells Following Transplantation: A Pre-Clinical Perspective - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC10888387/]
- 123. Cytohesin 1 regulates homing and engraftment of human hematopoietic stem and progenitor cells - PubMed [https://pubmed.ncbi.nlm.nih.gov/27899358/]
- 124. Dynamic rigidity changes enable rapid cell migration on ... [https://www.nature.com/articles/s41467-025-63854-9]
- 125. Animal models for cartilage repair [https://aber.apacsci.com/index.php/JBRHA/article/view/6359]
- 126. redefining rodent calvarial defect models for translational ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12577946/]
- 127. Drill-Hole Bone Defects in Animal Models of Bone Healing [https://www.researchprotocols.org/2022/7/e34887/]
- 128. Enhancement of in vitro and in vivo bone repair ... [https://www.nature.com/articles/s41598-025-99101-w]
- 129. A novel mouse model for full-thickness articular cartilage ... [https://www.sciencedirect.com/science/article/abs/pii/S0020138325003894]
- 130. Allogenic Bioengineered Cartilage Achieves Hyaline ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12125493/]
- 131. Reactive oxygen species regulate adipose-osteogenic ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12275293/]
- 132. bioengineered vascular conduit, autologous vein ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12593448/]
- 133. Novelties and limitations of tissue-engineered materials in ... [https://www.frontiersin.org/journals/cell-and-developmental-biology/articles/10.3389/fcell.2025.1603678/full]
- 134. Do adipose tissue-derived mesenchymal stem cells have ... [https://www.sciencedirect.com/science/article/pii/S1063458405001470]
- 135. Comparison of Osteogenic and Chondrogenic ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5960732/]